ISSN: 1839-9940
J Genomics 2021; 9:20-25. doi:10.7150/jgen.53678 This volume Cite
Research Paper
Draft genome sequences of three filamentous cyanobacteria isolated from brackish habitats
1. School of Geographical Sciences, Faculty of Science, University of Bristol, Bristol, BS8 1SS, United Kingdom.
2. Department of Marine Microbiology and Biogeochemistry, Royal Netherlands Institute for Sea Research, and Utrecht University, Den Hoorn, the Netherlands.
Abstract
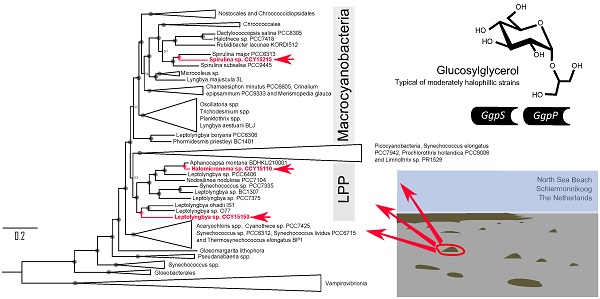
Brackish cyanobacterial genome sequences are relatively rare. Here, we report the 5.5 Mbp, 5.8 Mbp and 6.1 Mbp draft genomes of Spirulina sp. CCY15215, Leptolyngbya sp. CCY15150 and Halomicronema sp. CCY15110 isolated from coastal microbial mats on the North Sea beach of the island of Schiermonnikoog in the Netherlands. Large scale phylogenomic analyses reveal that Spirulina sp. CCY15215 is a large cell diameter cyanobacterium, whereas Leptolyngbya sp. CCY15150 and Halomicronema sp. CCY15110 are the first reported brackish genomes belonging to the LPP clade consisting primarily of Leptolyngbya, Plectonema and Phormidium spp. Further genome mining divulges that all new draft genomes contain, ggpS and ggpP, the genes responsible for synthesising glucosylglycerol (GG), a compatible solute found in moderately salt-tolerant cyanobacteria.
Keywords: Spirulina sp. CCY15215, Halomicronema sp. CCY15110, Leptolyngbya sp. CCY15150, cyanobacteria, genome, brackish

 Global reach, higher impact
Global reach, higher impact