ISSN: 1839-9940
J Genomics 2020; 8:21-24. doi:10.7150/jgen.42020 This volume Cite
Short Research Paper
Draft genome sequence of Deinococcus sp. KR-1, a potential strain for palladium-leaching
1. Research Institute for Sustainable Chemistry, National Institute of Advanced Industrial Science and Technology (AIST), 3-11-32 Kagamiyama, Higashi-Hiroshima, Hiroshima 739-0046, Japan
2. Department of Civil and Environmental Engineering, National Institute of Technology, Kure College, 2-2-11 Aga-minami, Kure, Hiroshima, 737-8506, Japan
3. Graduate School of Advanced Sciences of Matter, Hiroshima University, 1-3-1 Kagamiyama, Higashi-Hiroshima, Hiroshima 739-8530, Japan
4. Department of Civil Engineering, National Institute of Technology, Nagaoka College, Japan, 888, Nishi-Katagai, Nagaoka, Niigata 940-8532, Japan
Received 2019-11-12; Accepted 2020-1-2; Published 2020-2-10
Abstract
Strain KR-1 was isolated from pond water collected in Japan. Because this strain was capable of adsorbing palladium particles in sterilized water, strain KR-1 will be a useful biocatalyst for palladium-leaching from metal waste. Here we present a draft genome sequence of Deinococcus sp. KR-1, which consists of a total of 7 contigs containing 4,556,772 bp with a GC content of 70.0% and comprises 4,450 predicted coding sequences. Based on the 16S rRNA gene sequence analysis, strain KR-1 was identified as Deinococcus sp. KR-1.
Keywords: Deinococcus, Palladium-leaching, Draft genome sequencing, 16S rRNA, Phylogenetic tree
Introduction
Palladium is one of the platinum-group metals and is an important material used in several industries. For example, palladium is utilized in automobile catalytic converters, which are necessary to degrade harmful gases to harmless ones before their discharge into the atmosphere [1]. Palladium has a low melting point and is easily cast. For that reason, it is found on electronic circuit boards in many electronic devices, including televisions, computers and mobile phones [2]. Although the global demand of palladium is growing with the continuing development of technologies, the recovery rate of palladium from industrial products is low. However, in 2011, only 22% of the palladium was recovered from autocatalysts and jewelry scrap, and then its metal was reused [3]. Because electronic circuit boards include several types of rare metals, selective recovery of palladium entails multiple steps of pyro/hydrometallurgical processes, which increases the cost substantially [2]. Thus, establishment of a method for selective recovery of palladium would be highly desirable. Here we report the screening and isolation of a palladium-leaching bacterium, strain KR-1. For future in depth genomic studies and industrial applications of this strain, genome sequencing was carried out.
Materials and Methods
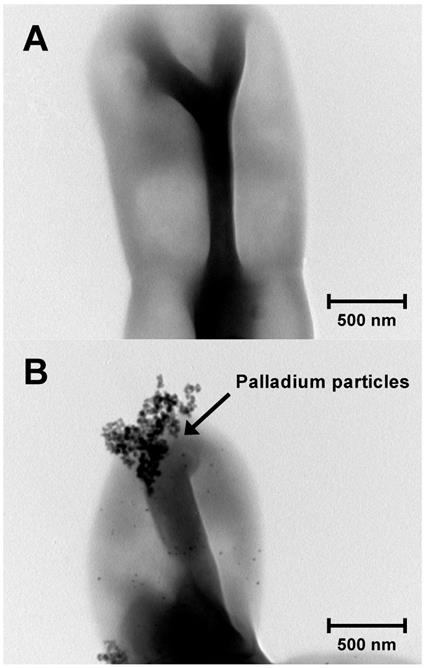
Pond water was collected from Kure city in Hiroshima prefecture, Japan. An R2A plate (pH 7.0; Nihon Pharmaceutical), which contained 9.7 g·L-1 K2HPO4, 3.2 g·L-1 KH2PO4, 0.05 g·L-1 peptone, 0.05 g·L-1 soluble starch, 0.05 g·L-1 yeast extract, 0.05 g·L-1glucose, 0.05 g·L-1 casamino acids, 0.03 g·L-1 sodium pyruvate and 0.005 g·L-1 MgSO4·7H2O was used for isolation. After 1 mL of a 0.0001% (v/v) diluted solution was inoculated onto the plate, the plate was incubated for 1 day at 30°C. Thereafter, several single colonies were re-streaked onto new plates at least three times to obtain pure colonies. After several purified strains were cultivated in R2A medium, the cells were collected by centrifugation and washed twice with sterile water. For palladium biosorption experiments, each washed strain was incubated for 1 h at room temperature in an oversaturated solution of palladium in water. To prepare samples for transmission electron microscopy (TEM), after hydrophilic treatment of copper grids (200 mesh; TED PELLA, USA), experimental samples (50 µL) were placed onto the treated copper grids as previously described [4]. TEM analysis was then carried out with a JEM-2011 Transmission Electron Microscope (JEOL, Tokyo, Japan) using an accelerating voltage of 200 kV. Qualitative analysis of metal particles carried out using an Energy Dispersive X-ray Spectrometer (JEOL) revealed that several isolates exhibited palladium biosorption.
We next determined a draft genome sequence for strain KR-1. A sample was prepared for genome sequencing by growing strain KR-1 aerobically overnight at 30°C in R2A medium. Genomic DNA was then extracted from the cultures using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The concentration and purity of the genomic DNA were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and a Quant-iT dsDNA BR assay kit (Invitrogen, Waltham, MA, USA). Sequencing of the genomic DNA was accomplished using MinION (Oxford Nanopore Technologies, Oxford, UK; Flow cell version R9 and rapid sequencing kit) and MiSeq (Illumina, San Diego, CA, USA) sequencers. Default parameters were used for all software used unless otherwise specified. Hybrid de novo assembly of the raw data was carried out using Unicycler ver.0.4.7 [5]. Genome annotation was performed using Prokka ver.1.14.0 [6].
TEM image of strain KR-1 (A) and strain KR-1 with palladium particles (B).
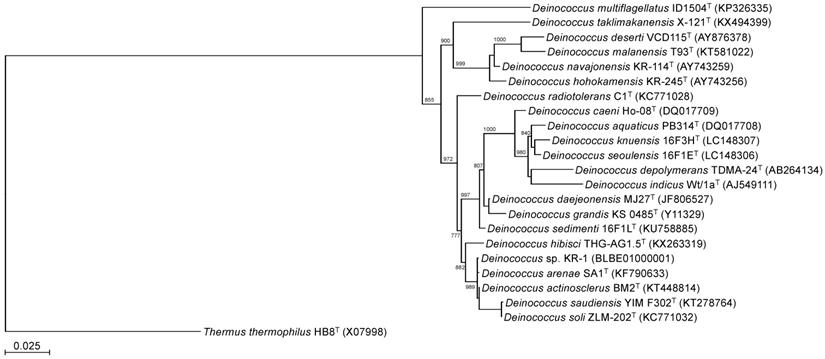
16S rRNA gene sequences of the related type strains were compared with reference sequences available in the GenBank/EMBL/DDBJ databases using BLAST. Construction of a maximum-likelihood tree was performed using FastTree2 [7, 8].
Results and Discussion
Although various biosorption methods for palladium-leaching are established [9, 10], most of the biosorption methods have not been applied in the industry [2]. The reasons are that their culture conditions are intricate and their growth is relatively slow. To establish a practicable biosorption method for palladium-leaching, we propose to apply an oligotroph with a faster growth rate. Because oligotroph grows under conditions of low levels of nutrients, our method may reduce the running cost. To obtain oligotrophic microorganisms, a diluted solution was prepared from pond water and then plated onto R2A plate. After incubation for 1 day at 30°C, several colonies were obtained. Subsequently, we carried out the biosorption experiment of palladium with several isolates. As the result, a few isolates showed palladium biosorption and the highest levels of biosorption were confirmed in strain K-1 (Fig. 1).
We next determined a draft genome sequence for strain KR-1. The raw data from the MinION and Miseq were 46,363 and 2,900,662 reads with 250-coverage. The genome sequence was 4,556,772 bp and the GC content was 70.0%. The assembly generated 7 contigs with an N50 contig size of 3,333,173 bp. Moreover, 4,450 predicted coding sequences were identified. In addition, 53 tRNA genes and 9 rRNA genes were detected using Prokka ver.1.14.0.
To identify the phylogeny of strain KR-1, a maximum likelihood tree based on the 16S rRNA gene sequences was constructed. In the resultant phylogenetic tree, strain KR-1 fell inside the cluster comprising members of the genus Deinococcus. Moreover, strain KR-1 exhibited similarities of 99.9 %, 99.9%, 98.5% and 98.8% to its closest relatives, D. arenae SA1T [11], D. actinosclerus BM2T [12], D. saudiensis YIM F302T [13] and D. soli ZLM-202T [14]. Thus, strain KR-1 was identified as Deinococcus sp. KR-1 (strain number: HUT-8138).
When genomic features of Deinococcus sp. KR-1, D. actinosclerus BM2T, D. actinosclerus SJTR and D. soli ZLM-202T were compared, numbers of tRNA and rRNA as well as the GC content were almost the same (Table 1). On the other hand, the coding sequence number of strain KR-1 was about 1.5 times higher than those of D. actinosclerus BM2T and D. soli ZLM-202T. This result suggested that strain KR-1 may have a unique pathway to grow in oligotrophic media. Although Desulfovibrio genus bacteria such as D. desulfurican D. fructosivorans and D. vulgaris have also the ability to adsorb palladium, the molecular mechanism of palladium-leaching is unclear [10, 15]. In this study, we determined the draft genome sequence of Deinococcus sp. KR-1, which enables to create the deletion mutants of cell wall protein. To elucidate the molecular mechanism of Deinococcus sp. KR-1, we are planning to examine palladium biosorption with several kinds of deletion mutants. The results will be described elsewhere as the next stage of our study.
Nucleotide Sequence Accession Number
The draft genome sequence of Deinococcus sp. KR-1 has been deposited in the DDBJ/EMBL/GeneBank under accession numbers BLBE01000001 to BLBE01000007. The raw sequence reads have been deposited in DDBJ under the BioProject number PRJDB8922 and BioSample number SAMD00192131.
Comparison of general genome features of strain KR-1 with the related type strains.
| Strain | Deinococcus sp. KR-1 (BLBE01000001-BLBE01000007) | D. actinosclerus BM2T (CP013910) | D. actinosclerus SJTR (CP029774) | D. soli ZLM-202T (CP011389) |
|---|---|---|---|---|
| Properties | ||||
| Genome length (bp) | 4,556,772 | 3,264,334 | 3,315,586 | 3,236,984 |
| GC content (%) | 70.0 | 70.6 | 70.6 | 70.2 |
| Contig numbers | 7 | 1 | 1 | 1 |
| Coding sequence numbers | 4,450 | 3,049 | 3727 | 2,944 |
| tRNA | 53 | 49 | 50 | 49 |
| rRNA | 9 | 9 | 12 | 9 |
Phylogenetic tree constructed from analysis of 16S rRNA gene sequences showing the relationships between strain KR-1 and its related type strains. The bar indicates a 0.05% nucleotide substitution rate. The tree was rooted using Thermus thermophilus HB8T as the outgroup.
Acknowledgements
We thank Dr. Makoto Maeda for technical assistance with TEM analysis. We are grateful to all members of the Bio-conversion Research Group at our Institute [Research Institute for Sustainable Chemistry, National Institute of Advanced Industrial Science and Technology (AIST)] for their technical assistance and valuable discussion.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Seo Y, Morimoto S. Analyzing platinum and palladium consumption and demand forecast in Japan. Resources. 2017;6:61
2. Jha MK, Lee J, Kim M, Jeong J, Kim B, Kumar V. Hydrometallurgical recovery/recycling of platinum by the leaching of spent catalysts: a review. Hydrometallurgy. 2013;133:23-32
3. Sarioğlan Ş. Recovery of palladium from spent activated carbon-supported palladium catalysts. Platinum Metals Rev. 2013;57:289-296
4. Kumagai A, Mizuno M, Kato N, Nozaki K, Togawa E, Yamanaka S, Okuda K, Saxena IM, Amano Y. Ultrafine cellulose fibers produced by Asaia bogorensis, an acetic acid bacterium. Biomacromolecules. 2011;12:2815-2821
5. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595
6. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068-2069
7. Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641-1650
8. Price MN, Dehal PS, Arkin AP. FastTree 2-Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490
9. Mack C, Wilhelmi B, Duncan JR, Burgess JE. Biosorption of precious metals. Biotechnol Adv. 2007;25:264-271
10. Vijayaraghavan K, Yun YS. Bacterial biosorbents and biosorption. Biotechnol Adv. 2008;26:266-291
11. Lee D, Cha S, Jang JH, Seo T. Deinococcus arenae sp. nov, a novel species isolated from sand in South Korea. Antonie Van Leeuwenhoek. 2016;109:1055-1062
12. Sun Joo E, Jin Lee J, Kang MS, Lim S, Jeong SW, Bit Kim E, Hwa Jeon S, Srinivasan S, Kyum Kim M. Deinococcus actinosclerus sp. nov, a novel bacterium isolated from soil of a rocky hillside. Int J Syst Evol Microbiol. 2016;66:1003-1008
13. Hussain F, Khan IU, Habib N, Xian WD, Hozzein WN, Zhang ZD, Zhi XY, Li WJ. Deinococcus saudiensis sp. nov, isolated from desert. Int J Syst Evol Microbiol. 2016;66:5106-5111
14. Cha S, Srinivasan S, Seo T, Kim MK. Deinococcus soli sp. nov, a gamma-radiation-resistant bacterium isolated from rice field soil. Curr Microbiol. 2014;68:777-783
15. Bennett JA, IMikheenko IP, Deplanche K, Shannon IJ, Wood J, Macaskie LE. Nanoparticles of palladium supported on bacterial biomass: New re-usable heterogeneous catalyst with comparable activity to homogeneous colloidal Pd in the Heck reaction. Appl Catal B. 2013;140-141:700-707
Author contact
![]() Corresponding author: Tel & Fax: +81-(0)823-73-8486; E-mail: z-kimurakure-nct.ac.jp
Corresponding author: Tel & Fax: +81-(0)823-73-8486; E-mail: z-kimurakure-nct.ac.jp

 Global reach, higher impact
Global reach, higher impact