ISSN: 1839-9940
J Genomics 2015; 3:51-56. doi:10.7150/jgen.11246 This volume Cite
Research Paper
Genetic Variability of MicroRNA Genes in 15 Animal Species
Department of Animal Science, Biotechnical Faculty, University of Ljubljana, Groblje 3, SI-1230 Domzale, Slovenia
Published 2015-2-15
Abstract
MicroRNAs (miRNA) are a class of non-coding RNAs important in posttranscriptional regulation of target genes. Previous studies have proven that genetic variability of miRNA genes (miR-SNP) has an impact on phenotypic variation and disease susceptibility in human, mice and some livestock species. MicroRNA gene polymorphisms could therefore represent biomarkers for phenotypic traits also in other animal species. We upgraded our previously developed tool miRNA SNiPer to the version 4.0 which enables the search of miRNA genetic variability in 15 animal genomes:
Keywords: cattle, livestock, microRNA, genetic polymorphisms, miRNA SNiPer
Implications
MicroRNAs have been identified as important regulators of gene expression and have been shown to be implicated in shaping phenotypic variability and disease development. In comparison to hundreds of expression studies, genetic variability residing within microRNA genes (miR-SNPs) has been much less explored; however they present an important pool of novel molecular biomarkers. We updated bioinformatics tool miRNA SNiPer 4.0 and performed genome-wide in silico screening of 15 animal genomes to collect miR-SNPs. We identified that currently cattle, human, drosophila and mouse genomes have the highest number of miR-SNPs. Additionally, the screening revealed several genomic hotspots for further functional analysis.
Introduction
MicroRNAs (miRNAs) are non-coding RNAs (ncRNAs), about 21 nucleotides in length, which bind to target mRNAs and post-transcriptionally regulate gene expression. By binding to different target gene regions they repress or activate translation [1, 2]. The key binding location for miRNA target recognition is the seed region, positioned 2-7 or -8 nucleotides from the 5'-end of the miRNA [3]. About half of miRNA genes reside within protein-coding host genes and if they are in the same orientation they share their transcriptional mechanisms [4].
Changes in miRNA expression and miRNA regulome polymorphisms have been associated with phenotypic traits and diseases [5, 6]. Therefore a systematic screening for miRNA gene variability could contribute to a development of new potential biomarkers associated to phenotype variability. In our previous study we collected polymorphic miRNA genes in livestock using miRNA SNiPer tool version 3.0 [7]. Due to new database updates it was necessary to update the tool and to perform a new genome-wide screening.
Material and methods
Online tool miRNA SNiPer 3.0 [7, 8] was upgraded to the version 4.0 which integrates data from four databases. MicroRNA gene location and genetic variability was obtained from the latest matching database assemblies of miRBase 21 and Ensembl Variation database, release 76. The Ensembl Variation database includes variation data from a variety of sources such as dbSNP. The locations of the seed miRNA regions were obtained from the TargetScan 6.2. The information related with commercial whole-genome SNP chips was obtained from SNPchiMp v.2 for five livestock species [9] (Additional File 1: Supplementary Tables S1, S2).
Results
In this study we updated the miRNA SNiPer tool 4.0 (http://www.integratomics-time.com/miRNA-SNiPer) and collected miRNA polymorphisms in 15 animal species. A catalog of multiple nucleotide polymorphisms (MNPs) within mature miRNA seed regions in cattle revealed several interesting genomic hotspots.
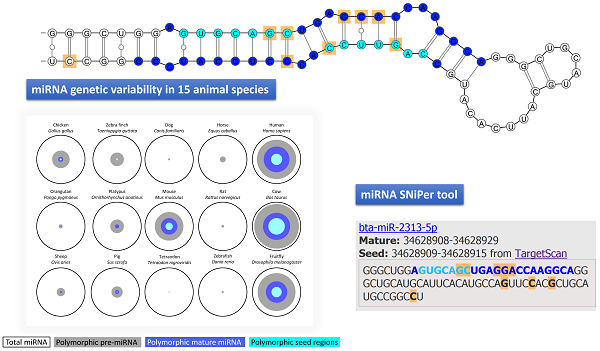
Among animal species with available miRBase and Ensembl Variation data 15 had matching database assemblies and were therefore included to the miRNA SNiPer tool: human, orangutan, cow, dog, horse, pig, sheep, mouse, rat, chicken, platypus, zebrafish, tetraodon, zebra finch, and fruitfly. In comparison to the version 3.0 dog and sheep genomes have been added and chimp, macaque and opossum were removed from the new tool update. The tool displays polymorphisms within pre-miRNA, mature and seed miRNA regions (Figure 1).
Output of the miRNA SNiPer tool displaying genetic variability of the bta-miR-764 gene. Polymorphisms are marked with the orange color. Details include information regarding the: 1. SNP location within the miRNA gene (seed region in light blue, mature region in dark blue and pre-miRNA region in black), 2. genomic location, 3. allele substitution, and 4. commercial whole-genome SNP chip name.
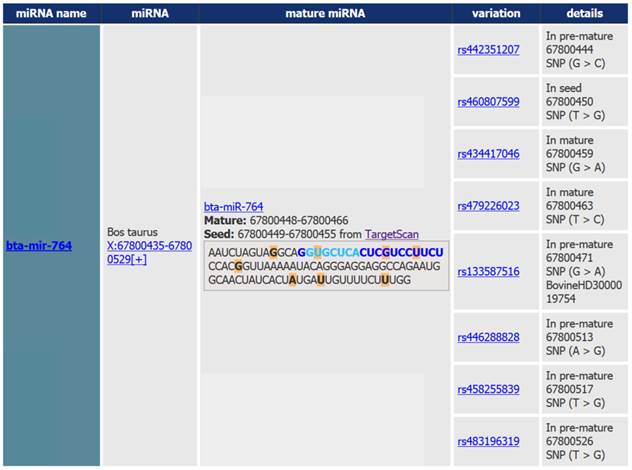
The updated version of the miRNA SNiPer tool was used for genome-wide screening of miRNA gene polymorphisms in 15 animal genomes. Based on current versions of genomic databases, miRNA genes are most polymorphic in cattle, followed by human, fruitfly, mouse, chicken, pig, horse, and sheep (Table 1, Figure 2). Screening revealed differences in the number of polymorphic miRNA genes between species, ranging from 3.8% (5/132) in tetraodon to 91.7% (741/808) in cattle.
Statistics of the data obtained using miRNA SNiPer version 4.0.
| Species in miRNA SNiPer 4.0 | Known miRNA genes | Polymorphic pre-miRNA regions | Polymorphic mature regions | Polymorphic mature seed regions | miRNA genes with seed MNP | DNA array data; SNPchiMp v.2 | miRNA genes with SNPs from DNA array |
|---|---|---|---|---|---|---|---|
| human | 1881 | 1532 | 991 | 437 | ND | no | 0 |
| orangutan | 642 | 52 | 14 | 4 | 0 | no | 0 |
| cattle | 808 | 741 | 452 | 278 | 61 | yes | 12 |
| horse | 715 | 86 | 15 | 8 | 0 | yes | 0 |
| pig | 383 | 89 | 30 | 7 | 0 | yes | 2 |
| sheep | 106 | 18 | 3 | 1 | 0 | yes | 2 |
| dog | 502 | 20 | 2 | 1 | 0 | no | 0 |
| mouse | 1193 | 707 | 406 | 176 | 10 | no | 0 |
| rat | 495 | 24 | 6 | 2 | 0 | no | 0 |
| chicken | 740 | 263 | 76 | 29 | 0 | yes | 26 |
| platypus | 396 | 11 | 3 | 0 | 0 | no | 0 |
| zebrafish | 346 | 8 | 0 | 0 | 0 | no | 0 |
| tetraodon | 132 | 5 | 0 | 0 | 0 | no | 0 |
| zebrafinch | 247 | 7 | 7 | 1 | 0 | no | 0 |
| fruitfly | 256 | 199 | 122 | 53 | 11 | no | 0 |
MNP = multiple nucleotide polymorphism; ND = not determined
Number of polymorphic pre-miRNA, mature miRNA and seed regions in 15 animal species included in the miRNA SNiPer tool 4.0.
Among collected miR-SNPs 46 are also present on commercial whole-genome SNP chips (Additional File 1: Supplementary Table S3). Most of the SNP array polymorphisms overlapped with pre-miRNA regions. Six polymorphisms in cattle and chicken also resided within miRNA mature and seed regions. Cattle miRNA gene bta-mir-1291 comprising seed SNP resided within two other genes: intron 9 of the protein-coding host gene KANSL2 and non-coding snoRNA gene SNORA2.
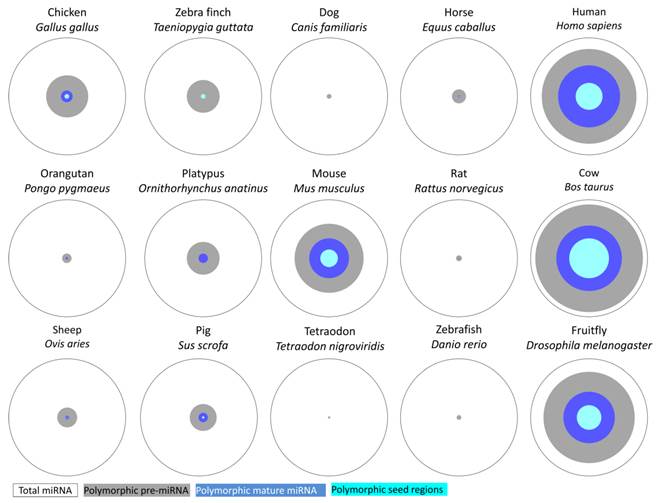
MicroRNA SNiPer 4.0 obtained significantly higher number of polymorphic miRNA genes than previous version. Using SNiPer 3.0 12% (92/766) of identified miRNA genes in cattle had polymorphic pre-miRNA regions and using SNiPer 4.0 91% (741/808) pre-miRNAs were polymorphic (Figure 3).
The most polymorphic pre-miRNA regions in cattle are bta-mir-212, bta-mir-763, and bta-mir-877, comprising 44 and 43 polymorphisms, respectively (Table 2). Out of 278 polymorphic bovine mature miRNA seed regions, there are 61 seed regions with consecutive polymorphisms; MNPs, which we have collected in a catalog (Additional File 1: Supplementary Table S4). MicroRNA bta-mir-346 had the longest region of consecutive polymorphisms; six out of seven nucleotides composing seed region are polymorphic. There is no information available regarding minor allele frequencies (MAF) of miR-346 seed polymorphisms; therefore this region should be sequenced. Out of 61 collected polymorphic miRNAs there are 38 intergenic and 23 located within introns of protein-coding genes. Some interesting genomic regions have been revealed, for example, an overlap of three genes (miRNA, snoRNA and protein-coding gene) has been identified on BTA14: bta-mir-1839, SCARNA15 and MROH1 (Figure 4). These overlapping regions are interesting for experimental analysis, since they could affect processing and function of miRNAs and snoRNAs.
Comparison of the results between the versions 3.0 and 4.0 of the miRNA SNiPer tool regarding the number of polymorphic pre-miRNA, mature miRNA and seed regions in cattle.
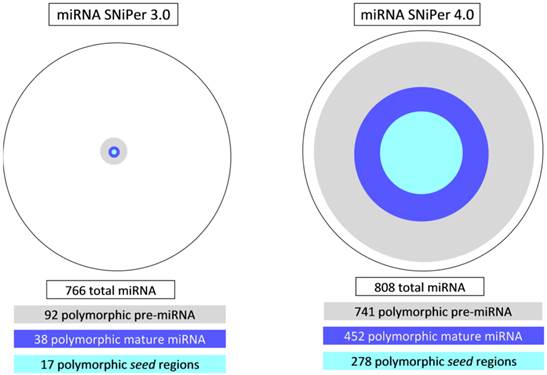
Overlap of three genes in cattle: miRNA gene bta-mir-1839, snoRNA gene SCARNA15, and protein-coding gene MROH1. Two consecutive SNPs are present within mature miRNA seed region. Dark blue - mature miRNA, light blue - seed region.

MicroRNA genes with the highest number of polymorphisms in cattle.
| miRNA | Number of pre-miRNA polymorphisms | Number of mature miRNA polymorphisms | Number of seed region polymorphisms |
|---|---|---|---|
| bta-miR-212 | 44 | 8 | 4 |
| bta-miR-763 | 43 | 9 | 5 |
| bta-miR-877 | 43 | 5 | 5 |
| bta-miR-1225-3p | 41 | 5 | 1 |
| bta-miR-125a | 32 | 5 | 2 |
| bta-miR-182 | 31 | 6 | 0 |
| bta-miR-138-1 | 28 | 5 | 1 |
| bta-miR-187 | 28 | 2 | 1 |
| bta-let-7b | 27 | 6 | 3 |
| bta-miR-132 | 26 | 2 | 3 |
| bta-miR-152 | 26 | 5 | 1 |
| bta-miR-346 | 24 | 3 | 6 |
Discussion
The amount of genome information is increasing rapidly; therefore regular updates of bioinformatics tools are essential. The current version of the miRNA SNiPer tool enables genome-wide analysis of miRNA polymorphisms in 15 animal genomes. The analysis will be possible also for turkey and cat when the miRBase and Ensembl assemblies will match. The difference in the number of miRNA-SNPs between species is most probably not due to a biological reason and lack of genetic variability in some species, but to still ongoing sequencing projects, differences in intensity of genome variability research and development of genomic resource databases.
High number of polymorphisms within miRNA genes especially within seed regions is surprising, since seed regions are believed to be highly conserved due to their key role in miRNA gene expression regulation. The seed region is responsible for target recognition and binding, therefore polymorphisms within this region could affect target recognition. Many among the collected polymorphisms are not validated and do not have available MAF value, therefore they could be results of sequencing errors.
Multiple nucleotide polymorphisms comprise more than one nucleotide, which means miRNA seed regions are no longer completely complementary to miRNA targets and therefore could cause gain/loss of a target. Because mature miRNA seed region is used for recognition of miRNA targets by perfect pairing with miRNA, MNPs have a greater chance of effect on miRNA function.
Since mature miRNA seed region is the most important region for miRNA-mRNA target recognition and accurate miRNA function, a catalog of miRNA seed region polymorphisms presents a valuable source of regions of potential interest for further functional analysis. Results of this study will be useful for further functional studies investigating the effects of miRNA gene polymorphisms on miRNA function, and particular on target recognition.
Supplementary Material
Additional File 1Supplementary Tables S1-S4
Acknowledgements
This work was supported by the Slovenian Research Agency (ARRS) through the Research programme (P4-0220) and project (Z4-6807).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33 doi:10.1016/j.cell.2009.01.002
2. Kunej T, Godnic I, Horvat S, Zorc M, Calin GA. Cross talk between microRNA and coding cancer genes. Cancer J. 2012;18:223-31 doi:10.1097/PPO.0b013e318258b771
3. Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15-20 doi:10.1016/j.cell.2004.12.035
4. Godnic I, Zorc M, Jevsinek Skok D, Calin GA, Horvat S, Dovc P. et al. Genome-wide and species-wide in silico screening for intragenic MicroRNAs in human, mouse and chicken. PLoS One. 2013;8:e65165. doi:10.1371/journal.pone.0065165
5. Obsteter J, Dovc P, Kunej T. Genetic variability of microRNA regulome in human. Molecular Genetics & Genomic Medicine. 2015:30-9
6. Georges M, Coppieters W, Charlier C. Polymorphic miRNA-mediated gene regulation: contribution to phenotypic variation and disease. Curr Opin Genet Dev. 2007;17:166-76 doi:10.1016/j.gde.2007.04.005
7. Jevsinek Skok D, Godnic I, Zorc M, Horvat S, Dovc P, Kovac M. et al. Genome-wide in silico screening for microRNA genetic variability in livestock species. Anim Genet. 2013;44:669-77 doi:10.1111/age.12072
8. Zorc M, Jevsinek Skok D, Godnic I, Calin GA, Horvat S, Jiang Z. et al. Catalog of MicroRNA Seed Polymorphisms in Vertebrates. PLoS One. 2012;7:e30737. doi:10.1371/journal.pone.0030737
9. Nicolazzi EL, Picciolini M, Strozzi F, Schnabel RD, Lawley C, Pirani A. et al. SNPchiMp: a database to disentangle the SNPchip jungle in bovine livestock. BMC Genomics. 2014;15:123. doi:10.1186/1471-2164-15-123
Author biography
 Dr. Minja Zorc is a postdoctoral fellow at the Department of Animal Science, Biotechnical Faculty, University of Ljubljana. She graduated from Computer and information science at the same University. Her main research interest is integration of genomics data and development of novel bioinformatics tools.
Dr. Minja Zorc is a postdoctoral fellow at the Department of Animal Science, Biotechnical Faculty, University of Ljubljana. She graduated from Computer and information science at the same University. Her main research interest is integration of genomics data and development of novel bioinformatics tools.
 Jana Obsteter obtained her BSc in Biotechnology from the Biotechnical Faculty at the University of Ljubljana. She is currently enrolled in MSc of Quantitative Genetics & Genome Analysis, Genetics of Human Complex Traits at the School of Biological Sciences, University of Edinburgh. Her primary research interest has been non-coding RNAs and she is currently enrolled in a project investigating genetic architecture of Type 2 diabetes and depression.
Jana Obsteter obtained her BSc in Biotechnology from the Biotechnical Faculty at the University of Ljubljana. She is currently enrolled in MSc of Quantitative Genetics & Genome Analysis, Genetics of Human Complex Traits at the School of Biological Sciences, University of Edinburgh. Her primary research interest has been non-coding RNAs and she is currently enrolled in a project investigating genetic architecture of Type 2 diabetes and depression.
 Dr. Peter Dovc is professor in Genetics and Animal Biotechnology at University of Ljubljana, Biotechnical Faculty and is heading Department of Animal Science. He has coauthored over 100 scientific papers in international journals and seven book chapters. His current research interests are identification of candidate genes for production traits and genetic diseases in domestic animals, biology of lactation, characterization of stem cells, conservation genetics and animal genome analysis.
Dr. Peter Dovc is professor in Genetics and Animal Biotechnology at University of Ljubljana, Biotechnical Faculty and is heading Department of Animal Science. He has coauthored over 100 scientific papers in international journals and seven book chapters. His current research interests are identification of candidate genes for production traits and genetic diseases in domestic animals, biology of lactation, characterization of stem cells, conservation genetics and animal genome analysis.
 Dr. Tanja Kunej is an associate professor at the Department of Animal Science, Biotechnical Faculty, University of Ljubljana, Slovenia. Dr. Kunej is co-author of 40 scientific publications, two book chapters and has received two US patent awards. Her main research interest is integration of genomics data in agriculture and biomedicine.
Dr. Tanja Kunej is an associate professor at the Department of Animal Science, Biotechnical Faculty, University of Ljubljana, Slovenia. Dr. Kunej is co-author of 40 scientific publications, two book chapters and has received two US patent awards. Her main research interest is integration of genomics data in agriculture and biomedicine.
![]() Corresponding author: Tanja Kunej, Chair of Genetics, animal biotechnology and immunology, Department of Animal Science, Biotechnical Faculty, University of Ljubljana, Groblje 3, Slovenia, Phone: +3861-320-3890; Fax: +3861 7217 888, Email: tanja.kunejuni-lj.si
Corresponding author: Tanja Kunej, Chair of Genetics, animal biotechnology and immunology, Department of Animal Science, Biotechnical Faculty, University of Ljubljana, Groblje 3, Slovenia, Phone: +3861-320-3890; Fax: +3861 7217 888, Email: tanja.kunejuni-lj.si

 Global reach, higher impact
Global reach, higher impact