ISSN: 1839-9940
J Genomics 2014; 2:54-58. doi:10.7150/jgen.7692 This volume Cite
Short Research Communication
Characterization of the Asian Citrus Psyllid Transcriptome
1. Genformatic, LLC, 6301 Highland Hills Drive Austin, TX 78731, USA.
2. Department of Biology, University of Nebraska at Omaha, Omaha, Nebraska 68182, USA.
3. Illumina, Inc., 25861 Industrial Blvd, Hayward, California 94545, USA.
4. Department of Plant Pathology and Plant-Microbe Biology, Cornell University, Ithaca New York 14853, USA.
5. Department of Entomology, Texas A&M University, 2475 TAMU College Station, Texas 77843, USA.
6. Heteroskedastic, Inc., Arvada, Colorado, USA.
7. Los Alamos National Laboratory Bioscience division B-6, MS M888 Los Alamos, New Mexico 87545, USA.
8. USDA Agricultural Research Service, 1503 South Providence Road, Columbia, Missouri 65203, USA.
9. School of Information, University South Florida, 4202 East Fowler Avenue, Tampa, FL 33260, USA.
10. University of Texas at Tyler, 3900 University Boulevard, Tyler, TX, 75799, USA.
11. USDA Agricultural Research Service, 2001 South Rock Road, Fort Pierce, FL 34945, USA.
12. Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Omaha, Nebraska 68182, USA.
* These authors contributed equally to this work.
Published 2014-2-10
Abstract
The Asian citrus psyllid, Diaphorina citri Kuwayama (Hemiptera: Psyllidae) is a vector for the causative agents of Huanglongbing, which threatens citrus production worldwide. This study reports and discusses the first D. citri transcriptomes, encompassing the three main life stages of D. citri, egg, nymph and adult. The transcriptomes were annotated using Gene Ontology (GO) and insecticide-related genes within each life stage were identified to aid the development of future D. citri insecticides. Transcriptome assemblies and other sequence data are available for download at the International Asian Citrus Psyllid Genome Consortium website [
Keywords: Asian Citrus Psyllid, Diaphorina citri Kuwayama
Results and Discussion
General Characteristics of the D. citri Transcriptomes
In total, 46,927,970 reads of 75 bp, 39,830,860 reads of 110 bp, and 50,248,212 reads of 100 bp were generated from the egg, nymph, and adult tissues, respectively, comprising 12.9 Gb of sequence, and were used to construct three de novo stage-specific transcriptomes (Table 1). The GC content of the transcriptomes was highly similar, ranging from 42.10% to 44.81%. Moreover, 99 to 100% of core eukaryotic genes had a detectable homolog in the stage-specific assemblies and between 84.3 and 88.2% of core genes had a detectable homolog whose alignment covered >80% of the core gene, indicating the transcriptomes were fairly complete.
Functional Annotation and Characterization of Diaphorina citri Transcripts
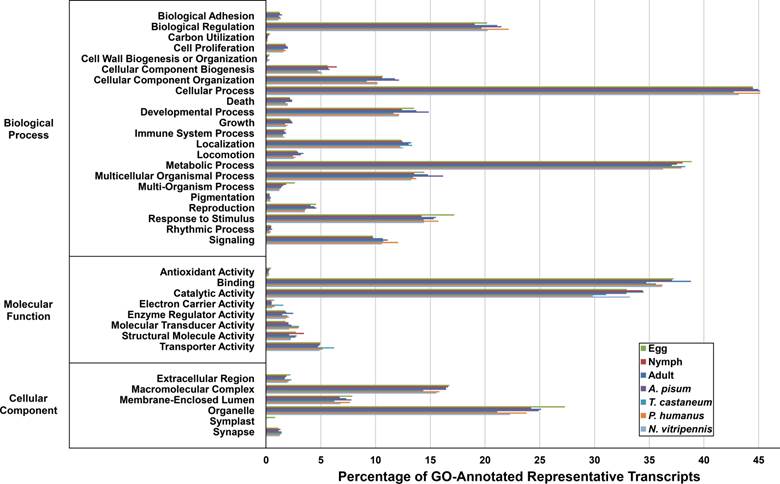
To review the putative functions of the D. citri transcripts and validate the completeness of the transcriptomes, GO analysis was performed against the three D. citri stages, plus four related organisms with complete transcriptome information. Of the total number of representative transcripts present within the egg, nymph and adult stages, 45.20% (26,546), 46.29% (26,651), and 40.00% (21,218) were assigned GO terms, respectively (Figure 1). Importantly, all four related organisms displayed this pattern, suggesting the completeness of the three stage transcriptomes.
Identification of Diaphorina citri Insecticide-Related Genes
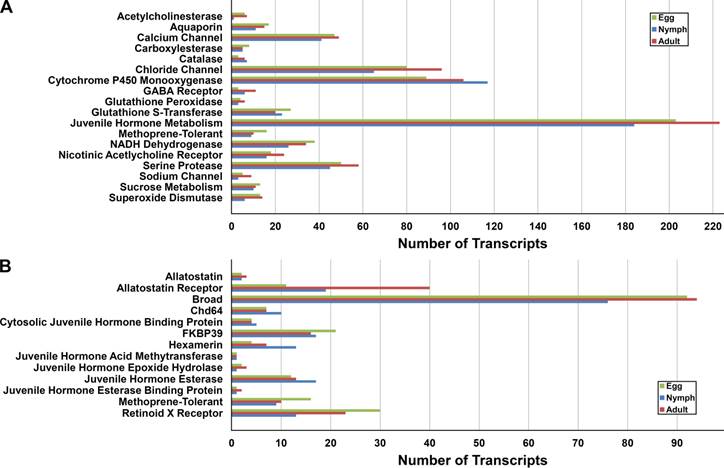
Insecticides are a pivotal component in controlling D. citri populations throughout the world, and therefore, it is essential to develop potent insecticides against a wide variety of molecular D. citri targets [1]. Insecticide targets and genes involved in detoxification and resistance were identified in the egg, nymph, and adult stages (Figure 2A). The largest and most complex group was 'Juvenile Hormone Metabolism.' Putative D. citri homologs of the major gene products involved in juvenile hormone metabolism exist within each life stage and exhibit a high degree of similarity to those genes of closely related organisms (Figure 2B and Table 2).
Statistics for Diaphorina citri transcriptome assemblies.
| Assembly | Stage | Total Number of Transcripts | Mean Contig Length | GC% | Core Eukaryotic Genes with Homologs | Core Eukaryotic Genes with Homologs (>80% coverage) | % of Transcripts Confirmed in Draft Genome Assembly | Putative Gene Fusions |
|---|---|---|---|---|---|---|---|---|
| Complete | Egg | 76,550 | 852 | 42.1 | 458/458 (100%) | 400/458 (87.3%) | 81.6% | 196 (0.256%) |
| - | Nymph | 69,233 | 756 | 44.81 | 456/458 (99.56%) | 404/458 (88.2%) | 81.0% | 146 (0.211%) |
| - | Adult | 62,450 | 691 | 42.96 | 458/458 (100%) | 386/458 (84.3%) | 81.0% | 158 (0.253%) |
| Representative* | Egg | 58,814 | 719 | 41.83 | 458/458 (100%) | 400/458 (87.3%) | 84.2% | 163 (0.278%) |
| - | Nymph | 57,635 | 632 | 44.27 | 456/458 (99.56%) | 403/458 (88.0%) | 79.0% | 123 (0.214%) |
| - | Adult | 53,117 | 595 | 42.67 | 458/458 (100%) | 386/458 (84.3%) | 88.7% | 147 (0.277%) |
Files containing the assembled transcripts for each of the D. citri life stages were analyzed using custom Perl scripts, and the total number of transcripts, mean contig length, %GC, number of core eukaryotic genes, % transcripts detectable in the genome assembly and number of putative gene fusions were determined. (See Methods section for details about how each was calculated.)
Juvenile hormone related genes identified in the Diaphorina citri transcriptome.
| Gene Name | NCBI Gene ID | FlyBase ID | Query ID | Query Length (bp) | Subject ID | Subject Species | E-Value |
|---|---|---|---|---|---|---|---|
| Allatostatin | 42947 | FBgn0015591 | diaci_nymph_66660000039047 | 692 | NP_001037036.1 | Bombyx mori | 3e-07 |
| Allatostatin Receptor | 44126 | FBgn0028961 | diaci_nymph_66660000026007 | 1547 | NP_524700.1 | Drosophila melanogaster | 0 |
| Broad | 44505 | FBgn0000210 | diaci_nymph_66660000033288 | 578 | |NP_726750.1 | Drosophila melanogaster | 0 |
| Chd64 | 38490 | FBgn0035499 | diaci_nymph_66660000018386 | 2227 | AAF47840.2 | Drosophila melanogaster | 1e-136 |
| Cytosolic Juvenile Hormone Binding Protein | 733092 | - | diaci_nymph_66660000009996 | 768 | NP_001037668.1 | Bombyx mori | 4e-142 |
| FKBP39 | 41860 | FBgn0013269 | diaci_nymph_66660000005954 | 2150 | CAA86996.1 | Drosophila melanogaster | 0 |
| Hexamerin | 660274 | - | diaci_nymph_66660000015491 | 2322 | NP_001164358.1 | Tribolium castaneum | 0 |
| Juvenile Hormone Acid Methytransferase | 34977 | FBgn0028841 | diaci_egg_55550000038216 | 1502 | EFA02917.1 | Tribolium castaneum | 3e-126 |
| Juvenile Hormone Epoxide Hydrolase | 251984 | FBgn0010053 | diaci_nymph_66660000003152 | 1779 | NP_001161902.1 | Tribolium castaneum | 2e-49 |
| Juvenile Hormone Esterase | 36780 | FBgn0010052 | diaci_adult_77770000038704 | 271 | NP_001180223.1 | Tribolium castaneum | 9e-84 |
| Juvenile Hormone Esterase Binding Protein | 37996 | FBgn0035088 | diaci_nymph_66660000032724 | 733 | NP_611989.1 | Drosophila melanogaster | 2e-18 |
| Methoprene-Tolerant | 32114 | FBgn0002723 | diaci_egg_55550000026684 | 389 | ABR25244.1 | Tribolium castaneum | 8e-39 |
| Retinoid X Receptor | 31165 | FBgn0003964 | diaci_nymph_66660000021934 | 2841 | AAF45707.1 | Drosophila melanogaster | 5e-93 |
Putative juvenile hormone related genes were identified using BLASTX to compare the representative D. citri transcripts from the egg, nymph and adult life stages to a custom BLAST database containing those genes of interest from closely related organisms. The top BLAST hits for the juvenile hormone related genes are shown.
Gene Ontology (GO) analysis of the representative transcripts present within the three life stages of Diaphorina citri. GO-terms were assigned to the representative transcripts of the three D. citri life stages and Acyrthosiphon pisum, Tribolium castaneum, Pediculus humanus and Nasonia vitripennis, the four organisms that exhibited the greatest number of top BLAST hits, using B2G4Pipe and then sorted into groups within three independent compartments using Blast2GO. The 'Cellular Process', 'Binding' and 'Organelle' groups contain the greatest number of representative transcripts within the 'Biological Processes', 'Molecular Function' and 'Cellular Component' domains, respectively. The representative transcriptomes of the four organisms and the egg, nymph and adult stages exhibited similar patterns, and thus indicates the completeness of the stage transcriptomes.
Genes related to insecticide targets, detoxification, and resistance within each of the three life stages of Diaphorina citri. Identification of putative D. citri insecticide-related genes was accomplished by comparing the representative transcripts from the three D. citri life stages to a custom protein database containing homologous genes of interest from closely related organisms using BLASTX. (A) D. citri predicted gene products of putative insecticide targets and predicted gene products putatively involved in detoxification and resistance were identified in all three life stages. (B) D. citri homologs of the major gene products involved in insect juvenile hormone metabolism are present within each life stage.
Methods
Growth, Sample Preparation, and Sequence Generation
Psyllids were field collected from citrus groves near USDA-ARS research station, 2001 South Rock Road, Fort Pierce, FL, 34945-3030; no specific permits were required. Psyllids were reared in 2' x 2' cages, temperature maintained at 26o C, lighting was dependent on natural day length throughout the year, and fed on Murraya paniculata for two years then Citrus macrophylla for the last two years. Tissue from one to four day old eggs, 3rd and 4th instar nymphs, and one day to one month old adults of mixed genders were collected, processed, and held at -80°C until RNA isolation.
Total RNA was isolated from whole egg, nymph, and adult tissues using Qiagen's RNeasy Mini Kit, per the manufacturer's instructions. The mRNA was purified using poly-T oligo-attached magnetic beads and converted into cDNA with random primers using the mRNA sequencing preparation kit from Illumina (part number 1004898). The cDNA was sequenced using an Illumina GAIIx sequencing system.
Transcriptome Assembly and Characterization
Velvet (v1.0.19) (k-mer of 47) and Oases (v0.1.19) were used to generate three stage-specific transcriptomes using reads from the egg, nymph or adult tissues, respectively [2,3]. Contigs with adapter sequence contamination were removed by alignment to Illumina adapter sequences using BWA [4].
The GC content and mean transcript length, were calculated using a custom Perl script to check completeness, transcriptomes were aligned with TBLASTN to sequences representing 458 core eukaryotic genes [5], and percent of core genes with at least one KOG family member that aligned to a transcript with an E-value < 1e-6 (optionally with alignment that covered 80% of KOG family member) was measured. To detect gene fusions, transcripts were aligned using BLASTN to 16,172 gene predictions produced using MAKER [134], and considered gene fusions if 1) the alignments between the transcript and two different MAKER gene predictions had an E-value < 1e-6 and percent identity >95%, 2) coordinates of the two alignments on the transcript overlapped by <10 nucleotides, 3) the two alignments together covered >95% of transcript sequence and 4) no alignment existed between transcript and MAKER model with an E-value <1e-6 and percent identity >95% that covered >95% of the transcript.
Generation of Representative Transcriptome Transcripts
To generate transcriptomes containing only unique transcripts for downstream analysis, by removing all repetitive, identical and near-identical transcripts, CD-HIT-EST was used with a sequence identity cut-off of 99% [6,7]. A loss of 23.17% (17,736), 16.75% (11,598), and 14.94% (9,333) of the total transcripts from the egg, nymph, and adult stages, respectively, was observed. Also, in order to compare D. citri to other previously sequenced Insecta spp., all NCBI RefSeq nucleotide records (retrieved August 12, 2012) were collected for Acyrthosiphon pisum, Tribolium castaneum, Pediculus humanus and Nasonia vitripennis totaling 17,675; 10,417; 10,775 and 12,927 sequences, respectively. In order to generate representative transcriptomes for each of these species, near-identical transcripts were removed using CD-HIT-EST using a cut-off value of 99% [6,7]. This resulted in a 4.64% (821), 4.09% (426), 0.47% (51) and 2.07% (267) loss of the total number of transcripts for A. pisum, T. castaneum, P. humanus and N. vitripennis, respectively.
Gene Ontology
To infer the putative function of the D. citri transcripts, BLASTX and an E-value threshold of ≤ 1e-5 was used to scan the representative transcriptomes against the NCBI non-redundant (nr) database (retrieved April 23, 2012). GO terms were assigned and were then categorized using the programs B2G4Pipe (version 2.5.0) and Blast2GO (version 2.5.1) using default settings, respectively, and the b2g_may12 GO database [8].
Identification of Insecticide-Related Genes
Insecticide-related genes present within the different life stages of D. citri were identified by using a combination of BLASTN, TBLAST, and BLASTX and an E-value threshold of ≤ 1e-5 to compare the representative transcriptomes to a custom BLAST database containing insecticide-related genes from A. pisum, Apis mellifera, Bombyx mori, Drosophila melanogaster, N. vitripennis, P. humanus, and T. castaneum (retrieved from NCBI on July 30, 2012).
Acknowledgements
This publication was supported by grants from the NIH National Center for Research Resources (5P20RR016469) and the National Institute for General Medical Science (8P20GM103427), USDA-ARS U.S. Horticultural Research Lab, Subtropical Insects Research Unit, Ft. Pierce, FL and a grant from the Citrus Research Board, Inc. (217 North Encina, P.O. Box 230, Visalia, CA. 93279). Its contents are the sole responsibility of the authors and do not necessarily represent the official views of NIH, NIGMS, USDA or CRB. Additional support was provided by FIRE and FUSE grants from the University of Nebraska at Omaha. This work utilized the Holland Computing Center of the University of Nebraska. We also acknowledge Biological Technicians Maria T. Gonzalez, Belkis Diego, Kathy Moulton, Ashley Voss, Carol Malone, PeiLing Li, USDA-ARS, U.S. Horticultural Research Laboratory, Fort Pierce, FL, 34945; Technicians: Chloe Hawkings, Evelien Van Ekert, Sulley K. Ben-Mahmoud, Erica R. Egan, Lindsay Shaffer, University of Florida-IFAS, IRREC, Fort Pierce, FL 34945; Tim Crouch, Associate Director, Networks and Operations, University of Texas at Tyler, TX; and Goutam Gupta, Los Alamos National Laboratory, Los Alamos, NM 87545. We acknowledge Angela Douglas and Xiangfeng Jing, Department of Entomology, Cornell University, NY, 14853 for the osmoregulatory analysis, and Dr. Xiomara Sinisterra, Science, Biotechnology Advisor, University of Texas at Tyler, TX, and Kevin Hodges.
Competing Interests
The authors have declared that no competing interests exist.
References
1. Hall DG, Ammar ED Richardson ML, Halbert SE. Asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae), vector of citrus huanglongbing disease. Entomologia Experimentalis et Applicata. 2012;146:207-223
2. Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086-1092
3. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821-829
4. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754-1760
5. Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061-1067
6. Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658-1659
7. Limin Fu, Beifang Niu, Zhengwei Zhu, Sitao Wu, Weizhong Li. CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics. 2012;28(23):3150-3152 doi: 10.1093/bioinformatics/bts565
8. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674-3676
Author contact
![]() Corresponding author: jreesecom (JR); wayne.hunterusda.gov (WBH).
Corresponding author: jreesecom (JR); wayne.hunterusda.gov (WBH).

 Global reach, higher impact
Global reach, higher impact