ISSN: 1839-9940
J Genomics 2022; 10:57-60. doi:10.7150/jgen.75652 This volume Cite
Research Paper
Draft Genomes of Six Wild Poisonous Mushrooms
1. Toxicology Center, National Institute of Health, Department of Medical Sciences, Ministry of Public Health, Nonthaburi 11000 Thailand.
2. Department of Biology, Faculty of Science, Mahasarakham University, Mahasarakham 44150, Thailand.
3. Department of Microbiology, Faculty of Science, Srinakharinwirot University, Bangkok 10110, Thailand.
4. Department of Biology, Faculty of Science, Srinakharinwirot University, Bangkok 10110, Thailand.
Received 2022-5-31; Accepted 2022-7-21; Published 2022-8-1
Abstract
Foodborne illnesses caused by wild mushroom poisoning occur globally and have led to food safety concerns. Here, we reported de novo genome assemblies of the six most commonly encountered toxic mushrooms in Thailand. These comprised Amanita brunneitoxicaria, Cantharocybe virosa, Chlorophyllum molybdites, Entoloma mastoideum, Pseudosperma sp. and Russula subnigricans. The nuclear genome sizes of these species ranged from 40 to 77 Mb, with the number of predicted genes ranging from 5,375 to 14,099. The mitogenome sizes varied from 41,555 to 78,907 bp. The resulting draft genomes of these poisonous mushrooms provide insights into toxin-related genes that may be used to establish genetic markers for monitoring mushroom poisoning outbreaks.
Keywords: Poisonous mushroom, De novo genome assembly, Toxin-related gene, Mitogenome
Introduction
Foraging wild mushrooms is a popular leisure activity. However, due to their morphological resemblance, poisonous mushrooms are often misidentified as edible species. Unintentional ingestion of toxic mushrooms can result in adverse poisoning effects, ranging from mild gastrointestinal symptoms to severe cytotoxic effects and death of patients [1-5]. Gastrointestinal irritant mushroom poisoning was most commonly encountered, followed by neurotoxic, cytotoxic, myotoxic and metabolic/endocrine toxicity mushroom poisoning [1]. This study revealed the first report of draft genomes of six common poisonous mushrooms in Thailand. These included Amanita brunneitoxicaria, Cantharocybe virosa, Chlorophyllum molybdites, Entoloma mastoideum, Pseudosperma sp. and Russula subnigricans. According to the classification of mushroom toxicity by White et al. [6], these mushrooms cause five major types of toxicity, including cytotoxicity (A. brunneitoxicaria, D346), neurotoxicity (Pseudosperma sp., D523), myotoxicity (R. subnigricans, D338), metabolic/endocrine toxicity (C. virosa, D287), gastrointestinal toxicity (C. molybdites, D392), and gastrointestinal toxicity coupled with neurotoxicity (E. mastoideum, D322). Although ingestion of most of these mushrooms resulted in mild poisoning symptoms [4,5,7], A. brunneitoxicaria and R. subnigricans caused fatal mushroom poisoning [1,3].
Materials and Methods
Mushroom samples were identified based on morphological and chemical characteristics, as well as ITS sequences [2-5]. Genomic DNA was extracted from fruiting bodies using the DNeasyTM Plant Mini Kit (QIAGEN, Germany). DNA concentrations of the samples were measured using fluorescence-based QubitTM quantitation assays (Qubit® 2.0 Fluorometer, Invitrogen, USA). Paired-end libraries (2 x 150 bp) were prepared using the TruSeq Nano DNA Kit (Illumina®, USA) and subsequently sequenced on the NovaSeq 6000 platform (Illumina®, USA). Paired-end reads were quality filtered using BBDuk [qtrim=rl, trimq=20, minlength=30], and normalized read coverage was obtained using BBNorm [target=40, mindepth=6] (http://jgi.doe.gov/data-and-tools/bb-tools/). The filtered paired-end reads were assembled using SPAdes V 3.15.2 with different k-mers [8]. Completeness evaluation of the final assemblies was conducted using BUSCO version 5.5.2 [9]. Eukaryotic gene prediction was carried out using AUGUSTUS version 3.4.0 [10,11] with parameters trained from Coprinopsis cinerea. The orthologous gene clusters were compared using OrthoVenn2 [12]. Fungal metabolite biosynthetic gene clusters were analyzed using antiSMASH 6.0 [13,14]. Circular mitochondrial contigs were aligned using BLAST against the Nr database (http://www.ncbi.nlm.nih.gov/blast).
Results and Discussion
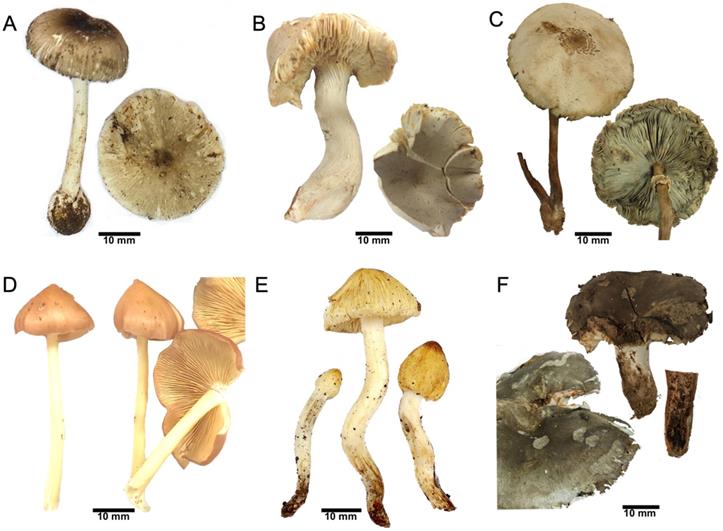
Six mushroom samples (Fig. 1) obtained from mushroom poisoning cases were left from food preparation. Detection of toxins revealed the presence of hepatotoxic amatoxins in A. brunneitoxicaria [3], and lethal cycloprop-2-ene carboxylic acid in R. subnigricans [15, 16].
Genome features of six poisonous mushrooms under study.
| Parameter | D346 | D287 | D392 | D322 | D523 | D338 |
|---|---|---|---|---|---|---|
| Genome coverage (x) | 104 | 109 | 115 | 111 | 106 | 97 |
| Number of contigs >1000 bp | 13,343 | 4,607 | 8,758 | 14,820 | 9,798 | 13,345 |
| Number of scaffolds >1000 bp | 12,482 | 4,450 | 8,073 | 13,983 | 8,970 | 12,650 |
| N50 of contigs (bp) | 9,588 | 32,238 | 9,063 | 4,695 | 9,933 | 7,564 |
| N50 of scaffolds (bp) | 10,450 | 34,423 | 10,221 | 5,274 | 11,467 | 8,248 |
| Longest scaffold length (kb) | 259.8 | 674.9 | 800.6 | 103.4 | 117.1 | 92.6 |
| Total scaffold length (Mb) | 77.1 | 52.0 | 49.0 | 51.8 | 55.7 | 64.2 |
| GC content (%) | 44.9 | 43.5 | 43.5 | 46.2 | 39.7 | 49.0 |
| Length of gap sequences (kb, ratio) | 87.4, 0.1% | 14.2, 0% | 32.8, 0.1% | 83.2, 0.2% | 56.7, 0.1% | 52.9, 0.1% |
| Number of genes predicted | 8,181 | 7,353 | 8,808 | 8,592 | 5,375 | 14,099 |
| Single-copy orthologs (%) | 92.6 | 95.7 | 92.5 | 85.9 | 89.4 | 84.0 |
| Number of biosynthetic genes encoding | ||||||
| Indole synthase | 1 | |||||
| Nonribosomal peptide synthetase (NRPS) | 2 | 2 | 2 | 1 | 2 | |
| Siderophore | 2 | 1 | ||||
| Terpene synthase | 7 | 8 | 16 | 5 | 6 | 8 |
| Type I polyketide synthase (PKS) | 1 | 1 | 1 | |||
| PKS-NRPS hybrid | 2 | |||||
| Mitochondrial genome (bp) | 62,094 | 41,555 | 78,907 | 47,464 | 66,949 | 66,439 |
| GC content (%) | 23.4 | 21.5 | 31.9 | 25 | 27.3 | 21.6 |
Mushroom samples. (A) A. brunneitoxicaria (D346), (B) C. virosa (D287), (C) C. molybdites (D392), (D) E. mastoideum (D322), (E) Pseudosperma sp. (D523) and (F) R. subnigricans (D338).
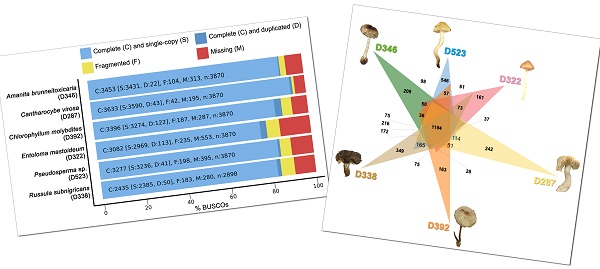
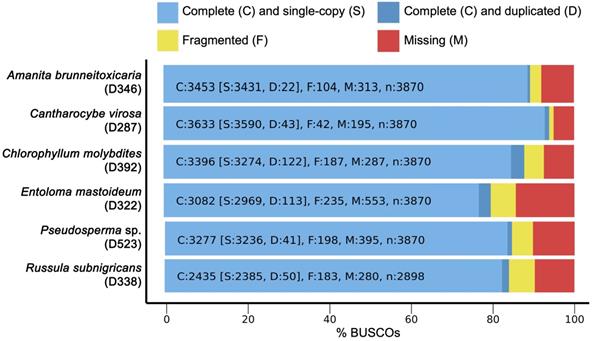
BUSCO completeness analysis for genome assemblies of six poisonous mushrooms.
(A) Venn diagram and (B) bar graph showing the number of gene families present in six poisonous mushrooms.
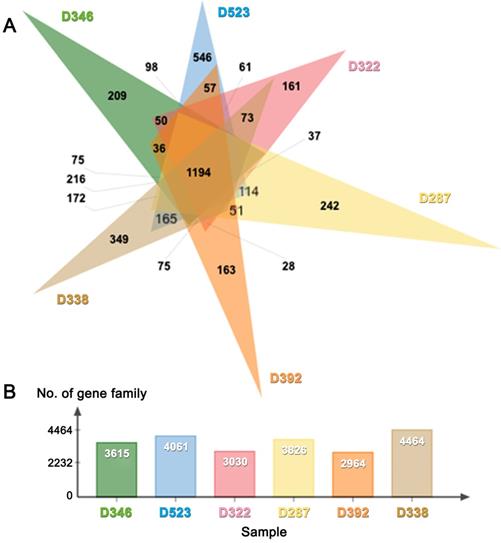
De novo draft genome assemblies revealed the median read depth ranging from 82x to 118x. The coverage of genomes ranged from 97x to 115x. The longest scaffold length (800,654 bp) was present in C. molybdites. Details of the genome features of the six poisonous mushrooms are shown in Table 1. The complete BUSCO scores ranged from 84% to 95.7% (Fig. 2). The number of protein-coding genes predicted ranged from 5,375 to 14,099. All six mushroom species possessed 6,796 gene clusters, including 6,045 orthologous gene clusters and 751 single-copy gene clusters. The Venn diagram showed 1,194 shared gene families (Fig. 3). Of the 68 biosynthetic gene clusters obtained, toxin-related genes have been the focus of the current study. The terpene synthase gene cluster was abundantly found in all mushroom genomes. This gene family corresponds to the largest group of secondary metabolite products in fungi [17]. The six mitogenomes ranged in size from 41,555 to 78,907 bp. For R. subnigricans, the size of mitogenome (66,439 bp) and GC content (21.6%) were similar to those reported in a previous study [18]. The data obtained from the resulting draft genomes of these poisonous mushrooms pave the way for the development of effective genome-based diagnostics for clinical application through in-depth genomic research.
Nucleotide Sequence Accession Numbers
Genome sequences were uploaded as BioProject PRJNA834754. The raw data were deposited in the NCBI/SRA database under the accession numbers SAMN28055954 (A. brunneitoxicaria, D346), SAMN28055957 (C. virosa, D287), SAMN28055958 (C. molybdites, D392), SAMN28055959 (E. mastoideum, D322), SAMN28055955 (Pseudosperma sp., D523) and SAMN28055956 (R. subnigricans, D338).
Acknowledgements
This work was financially supported by the National Institute of Health (NIH), Department of Medical Sciences, Ministry of Public Health and the Thailand Science Research and Innovation (TSRI) [grant number 164021/2022].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nooron N, Parnmen S, Sikaphana S, Leudang S, Uttawichai C, Polputpisatkul D. The situation of mushrooms food poisoning in Thailand: symptoms and common species list. Thai J Toxicol. 2020;35(2):58-69
2. Parnmen S, Sikaphan S, Leudang S, Boonpratuang T, Rangsiruji A, Naksuwankul K. Molecular identification of poisonous mushrooms using nuclear ITS region and peptide toxins: a retrospective study on fatal cases in Thailand. J Toxicol Sci. 2016;41(1):65-76
3. Ramchiun S, Sikaphan S, Leudang S, Polputpisatkul D, Nantachaiphong N, Khaentaw T. et al. Molecular characterization and liquid chromatography-mass spectrometric multiple reaction monitoring-based detection in case of suspected phalloides syndrome poisoning. J Assoc Med Sci. 2019;52(1):48-55
4. Parnmen S, Nooron N, Leudang S, Sikaphan S, Polputpisatkul D, Rangsiruji A. Phylogenetic evidence revealed Cantharocybe virosa (Agaricales, Hygrophoraceae) as a new clinical record for gastrointestinal mushroom poisoning in Thailand. Toxicol Res. 2020;36(3):239-248
5. Parnmen S, Nooron N, Leudang S, Sikaphan S, Polputpisatkul D, Pringsulaka O. et al. Foodborne illness caused by muscarine-containing mushrooms and identification of mushroom remnants using phylogenetics and LC-MS/MS. Food Control. 2021;128:108182
6. White J, Weinstein SA, De Haro L, Bédry R, Schaper A, Rumack BH. et al. Mushroom poisoning: A proposed new clinical classification. Toxicon. 2019;157:53-65
7. Leudang S, Sikaphan S, Parnmen S, Nantachaiphong N, Polputpisatkul D, Ramchiun S. et al. DNA-based identification of gastrointestinal irritant mushrooms in the genus Chlorophyllum: A food poisoning case in Thailand. J Heal Res. 2017;31(1):41-49
8. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De Novo Assembler. Curr Protoc Bioinforma. 2020;70(1):e102
9. Simão FA, Waterhouse RM, Ioannidis P, Kriventseva E V, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210-3212
10. Keller O, Kollmar M, Stanke M, Waack S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 2011;27(6):757-763
11. Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:ii215-225
12. Xu L, Dong Z, Fang L, Luo Y, Wei Z, Guo H. et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47(W1):W52-W58
13. Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH. et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49(W1):W29-W35
14. Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA. et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339-W346
15. Matsuura M, Saikawa Y, Inui K, Nakae K, Igarashi M, Hashimoto K. et al. Identification of the toxic trigger in mushroom poisoning. Nat Chem Biol. 2009;5(7):465-467
16. Matsuura M, Kato S, Saikawa Y, Nakata M, Hashimoto K. Identification of Cyclopropylacetyl-(R)-carnitine, a unique chemical marker of the fatally toxic mushroom Russula subnigricans. Chem Pharm Bull (Tokyo). 2016;64(6):602-608
17. Wang Q, Cao R, Zhang Y, Qi P, Wang L, Fang S. Biosynthesis and regulation of terpenoids from basidiomycetes: exploration of new research. AMB Express. 2021;11:150
18. Yu F, Zhang Y, Liang J. The mitochondrial genome of a wild toxic mushroom, Russula subnigricans. Mitochondrial DNA Part B. 2019;4(2):4126-4127
Author contact
![]() Corresponding author: Tel & Fax: +66-26495000; E-mail: achariyaswu.ac.th.
Corresponding author: Tel & Fax: +66-26495000; E-mail: achariyaswu.ac.th.

 Global reach, higher impact
Global reach, higher impact