ISSN: 1839-9940
J Genomics 2019; 7:11-13. doi:10.7150/jgen.32210 This volume Cite
Short Research Communication
Draft Genome Sequence of the Oleaginous Yeast Apiotrichum porosum (syn. Trichosporon porosum) DSM 27194
Institute of Process engineering in Life Science 2: Technical Biology, Karlsruhe Institute of Technology, Germany
* Olga Gorte and Habibu Aliyu are co-first authors.
Received 2018-12-12; Accepted 2019-1-22; Published 2019-1-29
Abstract
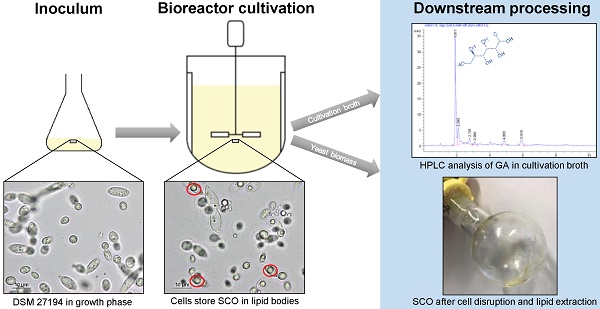
Here, we present the draft genome sequence of Apiotrichum porosum DSM 27194 generated on PacBio platform. Characterization of this oleaginous yeast originally collected from the grassland in Karlsruhe Germany, revealed potential for its utilization as a source of single cell oil (SCO) and gluconic acid (GA). The availability of the genome sequence provides a valuable resource for the elucidation of the genetic processes determining SCO and GA biosynthesis.
Keywords: Apiotrichum porosum DSM 27194, draft genome, gluconic acid, oleaginous yeast and single cell oil.
Introduction
Apiotrichum porosum DSM 27194 was observed in a bioreactor fermentation on glucose as a simultaneous single cell oil (SCO) and gluconic acid (GA) producer, with yields of 17.0 g/L SCO and 12 g/L GA. Fermentation, using xylose as carbon source, yielded of 13.9 g/L SCO [1]. This oleaginous yeast was isolated from a grassland in Karlsruhe, identified as Trichosporon porosum via ITS region sequencing and finally deposited to the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) as Trichosporon porosum DSM 27194 [1]. Due to the discovered properties, this basidiomycete species can be an interesting candidate for a wide range of biotechnological processes. Especially, by facing the pending economic challenges, namely the foreseeable depletion of crude oil, the highly controversial “food-or-fuel” debatte, overfishing of the oceans and the urgent need for the reduction of greenhouse gas emissions, microbial SCO can be used as potential alternatives for crude, plant, and fish oil [2]. GA is regarded as a bulk chemical in the textile, pharmaceutical, and construction industries and is highly used for food manufacturing [3].
The whole genome sequencing of A. porosum DSM 27194 provides the genetic information for its yet unknown SCO and GA metabolic pathways. In addition, this data will enable new possibilities for phylogenetic and comparative genomics investigations.
Materials and Methods
Genomic DNA was extracted from A. porosum DSM 27194 after cultivating in a mineral salt medium described in [1], containing a phosphate buffer system at pH 5 (8.99 g/L KH2PO4 and 0.12 g/L Na2HPO4 x 2 H2O), 0.1 g/L sodium citrate x 2 H2O, 0.1 g/L yeast extract, 0.2 g/L MgSO4 x 7 H2O, 4.72 g/L (NH4)2SO4 . After autoclaving 2 % (v/v) of sterile trace elements solution with 4 g/L CaCl2 x 2 H2O, 0.55 g/L FeSO4 x 7 H2O, 0.475 g/L citric acid, 0.1 g/L ZnSO4 x 7 H2O, 0.076 g/L MnSO4 x H2O, 100 μl/L 18 M H2SO4 and 2 % of sterile salt solution containing 20 g/L MgSO4 x 7 H2O, 10 g/L yeast extract was added. In addition, 50 g/L glucose was used as carbon source. The cells were cultivated in three replicate cultures in conical shake flasks at 25 °C and 130 rpm until early logarithmic growth phase.
Core genome phylogeny of Apiotrichum porosum DSM 27194, 16 related Trichosporonales and Cryptococcus amylolentus CBS 6039T (outgroup). The maximum likelihood (ML) phylogeny was inferred from the alignment of 132-consensus single-copy proteins using PhyML based the LG+G+I+F substitution model determined using SMS. The ML was generate with confidence values based on 1000 bootstrap replicates.
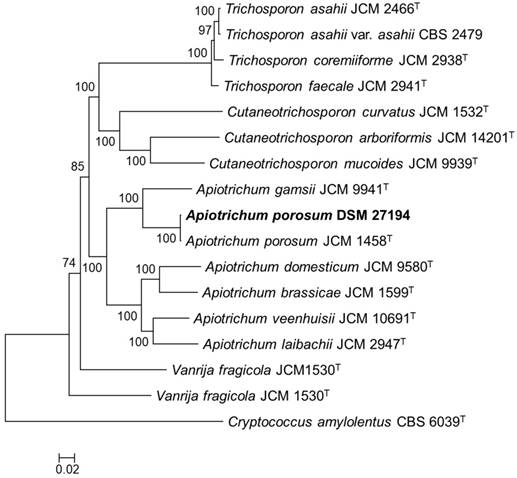
The genomic DNA of Apiotrichum porosum DSM 27194 was sequenced with PacBio long reads chemistry at the Microsynth AG (Balgach, Switzerland) and GATC Biotech AG (Konstanz, Germany). The reads, comprising of 1,465,786,355 bases were first assembled using Canu v1.7.1 [4] and the pre-assembled contigs polished with arrow v2.3.2 [5]. Structural and function annotation of the polished assembly was conducted using Funannotate pipeline v1.5.0-8f86f8c (https://github.com/nextgenusfs/funannotate). Completeness of the draft genome of DSM 27194 was assessed using BUSCO v2.0 [6]. For genome-wide phylogeny reconstruction, additional genome sequences of sixteen validly described members of the Trichosporonales were annotated using Funannotate pipeline and consensus single-copy protein sequences identified based on OrthoFinder [7] and BUSCO (fungi_odb9) among the predicted proteins of the seventeen genomes. The single-copy proteins were aligned using T-Coffee [8], concatenated and GBlocks [9] trimmed. The concatenated protein alignment was used to generate a maximum likelihood phylogeny with confidence values based on 1000 bootstrap replicates using PhyML v20110919 under the LG+G+I+F substitution model predicted using SMS [10].
Results and Discussion
The assembled draft genome sequence of DSM 27194 consists of 32 contigs totalling 25,479,456 bp (mean coverage: 56.4x) with a mean G+C content of 59.15%. Genome sizes and mean G+C contents ranges among available genomes of Apiotrichum spp are 31,617,680 - 23,647,732 bp and 61.14 - 56.47%, respectively. The sizes of the largest and N50 contigs were 3,134,003 and 1,376,709 bp, respectively. Annotation of the genome yielded 9,729 genes of which 9,153 code for proteins and 576 constitute tRNA genes. Evaluation of the predicted proteins with fungi_odb9 and basidiomycota_odb9 indicated a genome completeness of 97.9% and 96.7%, respectively. This compares closely with fungi_odb9 completeness values of between 96.90 and 90.60% estimated for related members of the genus Apiotrichum. Further evaluation of the predicted proteins revealed 570 putative carbohydrate-activating enzymes (CAZYmes), 575 secretome and 265 MEROPS associated predictions.
To infer the phylogenetic relation of DSM 27194, maximum likelihood phylogeny was constructed based on alignment of 132-consensus single-copy proteins, comprising 45,269 amino acids in length, of seventeen isolates selected from the genus Apiotrichum and 3 closely related genera in the family Trichosporonaceae (Figure 1). Consistent with earlier identification of DSM 27194, based on the sequence of the ITS region, the whole-genome phylogeny showed that DSM 27194 cluster with Apiotrichum porosum (syn. Trichosporon porosum) JCM 1458T. However, genomes of the two isolates differ slightly in terms of mean G+C contents (DSM 27194: 59.10% and JCM 1458T: 58.52%) and number of unique proteins which stand at 621 in DSM 27194 and 705 in JCM 1458T.
Evaluation of the DSM 27194 genome revealed that it encodes the key enzymes that have been demonstrated to play a central role in SCO production [11, 12]. These include putative AMP deaminase (EHS24_003651), isocitrate dehydrogenase (EHS24_007601, EHS24_002540 and EHS24_002733), malic enzyme (EHS24_008089), ATP citrate lyase (EHS24_004735), acetyl-CoA carboxylase (EHS24_004089) and fatty acid synthase (EHS24_002351, EHS24_002352). The genome also encodes multiple copies of glucose oxidase (EHS24_007400, EHS24_008706, EHS24_008979, EHS24_009251, EHS24_009561), which is mandatory for the first step in the oxidation of β-D-glucose to GA. The second step of the reaction can occur spontaneously [13].
Subsequent in-depth comparative genomics strategy and analytical methods will provide insight into the genetic determinants high lipid yields reported in A. porosum DSM 27194.
Nucleotide Sequence Accession Number
The whole genome sequence of Apiotrichum porosum DSM 27194 has been deposited at DDBJ/EMBL/Genbank under the accession number RSCE00000000. The version described in this paper is the first version.
Acknowledgements
Bioeconomy International BMBF (grant #031B0452) supported OG. HA acknowledges funding from Alexander von Humboldt Foundation. The authors wish to acknowledge the use of computational resources provided by Siemens. The authors wish to acknowledge support by Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of Karlsruhe Institute of Technology.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Schulze I, Hansen S, Großhans S, Rudszuck T, Ochsenreither K, Syldatk C. et al. Characterization of newly isolated oleaginous yeasts - Cryptococcus podzolicus, Trichosporon porosum and Pichia segobiensis. AMB Express. 2014;4:24
2. Ochsenreither K, Glück C, Stressler T, Fischer L, Syldatk C. Production Strategies and Applications of Microbial Single Cell Oils. Frontiers in microbiology. 2016;7:1539 -
3. Singh OV, Kumar R. Biotechnological production of gluconic acid: future implications. Appl Microbiol Biotechnol. 2007;75:713-22
4. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722-36
5. Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563
6. Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G. et al. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol Biol Evol. 2018;35:543-8
7. Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology. 2015;16:157
8. Taly J-F, Magis C, Bussotti G, Chang J-M, Di Tommaso P, Erb I. et al. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. nature protocols. 2011;6:1669-82
9. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540-52
10. Lefort V, Longueville J-E, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34:2422-4
11. Shen Q, Chen Y, Jin D, Lin H, Wang Q, Zhao Y-H. Comparative genome analysis of the oleaginous yeast Trichosporon fermentans reveals its potential applications in lipid accumulation. Microbiol Res. 2016;192:203-10
12. Adrio JL. Oleaginous yeasts: Promising platforms for the production of oleochemicals and biofuels. Biotechnol Bioeng. 2017;114:1915-20
13. Ramachandran S, Fontanille P, Pandey A, Larroche C. Gluconic acid: Properties, applications and microbial production. Food Technology & Biotechnology. 2006:44
Author contact
![]() Corresponding authors: Habibu Aliyu. Mailing address: habibu.aliyukit.edu. Telephone: +49 721 608-42125. Katrin Ochsenreither. Mailing address: katrin.ochsenreitheredu. Telephone: +49 721 608-46478.
Corresponding authors: Habibu Aliyu. Mailing address: habibu.aliyukit.edu. Telephone: +49 721 608-42125. Katrin Ochsenreither. Mailing address: katrin.ochsenreitheredu. Telephone: +49 721 608-46478.

 Global reach, higher impact
Global reach, higher impact