ISSN: 1839-9940
J Genomics 2017; 5:32-35. doi:10.7150/jgen.19407 This volume Cite
Short Research Communication
Complete genome sequence and functional study of the fibrinolytic enzyme-producing bacterium Leuconostoc holzapfelii 5H4, a silage probiotic
1. Division of Applied Life Science (BK21 Plus) and Institute of Agriculture & Life Science, Gyeongsang National University, Jinju 52828, Korea;
2. Central Area Crop Breeding Division, NICS, RDA, Suwon 16429, Korea.
Published 2017-3-9
Abstract
To process silage, rye is usually removed before the heading stage but the rye biomass increased up to 30% after the heading stage. However, after the heading stage, lignification rapidly accelerated and it resulted in a poor NDF digestibility problem. This has led to a demand for a strong fibrinolytic enzyme-producing probiotic for rye silage. The Gram-positive Leuconostoc holzapfelii 5H4 was selected as a fibrinolytic enzyme-producing probiotic to overcome lignification of rye silage. The L. holzapfelii 5H4 has a relatively small circular chromosome (1,885,398 bp), but the strain has one cellulase, two xylanase, and five esterase in its genome sequence. All fibrinolytic enzyme genes were relatively highly expressed compared to housekeeping genes, and this was confirmed by qRT-PCR. In this study, we report the complete genome sequence of the bacterium so that fibrinolytic enzyme production and its fibrinolytic activity mechanism are better understood.
Keywords: Complete genome sequencing, Lignification, NDF digestibility
Genome Announcement
The genus Leuconostoc belongs to the order Lactobacillales, an order of Gram-positive bacteria within the phylum Firmicutes. Like other lactic acid bacteria, Leuconostoc sp. strains produce lactic acid as the major metabolite of sugar fermentation. They are used in the production of fermented products such as cheese, butter, buttermilk, kefir, sourdough and kimchi. At the time of writing the genus Leuconostoc comprised 14 species. Recently, several novel Leuconostoc species have been described, originating from different types of food. These species include Leuconostoc kimchii from kimchi, a Korean vegetable product (1), Leuconostoc gasicomitatum from marinated broiler meat strips (2), Leuconostoc durionis from tempoyak, fermented durian (3), and Leuconostoc ficulneum and Leuconostoc pseudoficulneum from ripe figs (4, 5).
Leuconostoc holzapfelii strain 5H4 was isolated from rye silage in South Korea. Ferulate esterases cleave the ester linkage, which increases the digestibility of silage (6). Donaghy et al. (7) have reported that several subspecies of Lactobacillus produce ferulate esterases. Although similar to increase the fibrinolytic activity of rye silage, based on genetic and biological analyses, L. holzapfelii 5H4 possesses different characteristics than other LAB (Kim et al. unpublished data). Therefore to understand this bacterium, including its fibrinolytic activity, its complete genome was sequenced in this study.
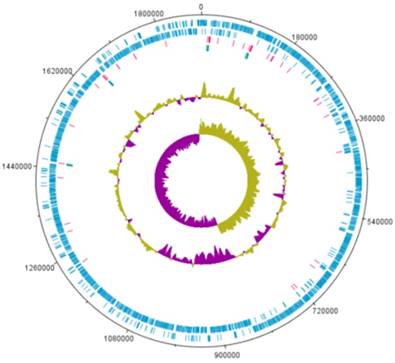
A complete genome sequence of L. holzapfelii strain 5H4 was determined with PacBio sequencing. The genome of L. holzapfelii strain 5H4 comprises a single circular chromosome (1,885,398bp) (Table 1, Fig. 1). The G+C contents of this component is 38.7%. This strain carries 70 tRNAs, and 12 rRNAs. The chromosome size of L. holzapfelii strain 5H4 was similar to a previously reported Leuconostoc spp. genome (8). Of the 1816 identified genes, 1227 were classified into different functional categories based on the subsystem category distribution (Table 2). Most of the genes in L. holzapfelii 5H4 were associated with functions such as carbohydrate, amino acids and derivatives, DNA metabolism, and protein metabolism. Genes encoding esterase, cellulase and xylanas protein were found in the genome of L. holzapfelii strain 5H4 (Table 3). Only one gene encoding cellulase protein was present on the chromosome of L. holzapfelli strain 5H4. Two gene encoding xylanase proteins were present on the chromosome of L. holzapfelli strain 5H4. Five gene encoding esterase proteins were revealed in L. holzapfelli strain 5H4 genome.
Genome features of Leuconostoc holzapfelii strain 5H4.
| Features | Chromosome |
|---|---|
| Genome size | 1,885,398 bp |
| G C content (%) | 38.7 |
| CDS | 1,839 |
| tRNA genes | 70 |
| rRNA genes | 12 |
Genome map of L. holzapfelii strain 5H4.
Functional categories of Leuconostoc holzapfelii strain 5H4.
| Functional description | Number of genes |
|---|---|
| Cofactors, Vitamins, Prosthetic Groups, Pigments | 97 |
| Cell Wall and Capsule | 89 |
| Virulence, Disease and Defense | 26 |
| Potassium metabolism | 8 |
| Miscellaneous | 18 |
| Phages, Prophages, Transposable elements, Plasmids | 13 |
| Membrane Transport | 43 |
| RNA metabolism | 73 |
| Nucleosides and Nucleotides | 88 |
| Protein Metabolism | 183 |
| Cell Division and Cell cycle | 40 |
| Regulation and cell signaling | 17 |
| DNA metabolism | 104 |
| Fatty Acid, Lipids, and Isoprenoids | 62 |
| Nitrogen Metabolism | 4 |
| Respiration | 13 |
| Stress response | 29 |
| Metabolism of Aromatic Compounds | 3 |
| Amino Acids and Derivatives | 132 |
| Sulfur Metabolism | 6 |
| Phosphorus Metabolism | 32 |
| Carbohydrates | 147 |
List of fibrinolytic enzyme-related genes of Leuconostoc holzapfelii 5H4
| Gene | start | stop | Length (bp) | Function |
|---|---|---|---|---|
| Cellulase | 1372885 | 1371809 | 1077 | Cellulase M |
| Xylanase1 | 1560810 | 1561589 | 1410 | Xylanase |
| Xylanase2 | 1561707 | 1562669 | 963 | Xylanase |
| Esterase1 | 1776728 | 1775682 | 1047 | Esterase/lipase |
| Esterase2 | 1822828 | 1823628 | 801 | probable lipase/esterase |
| Esterase3 | 1823659 | 1824462 | 804 | putative esterase |
| Esterase4 | 1824662 | 1825327 | 666 | Esterase/lipase |
| Esterase5 | 1839055 | 1839957 | 903 | Esterase/lipase |
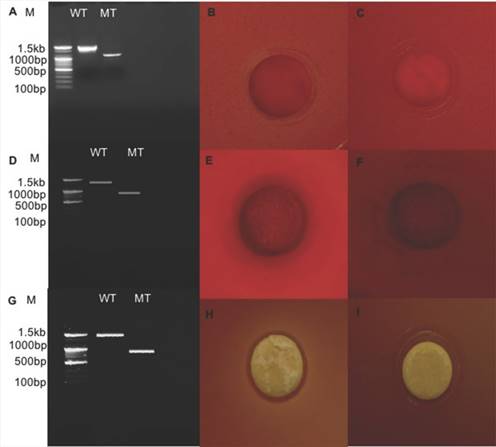
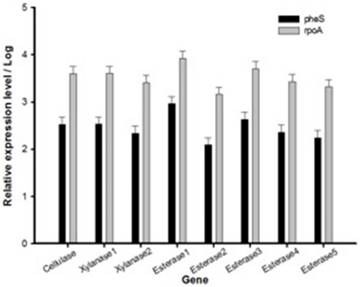
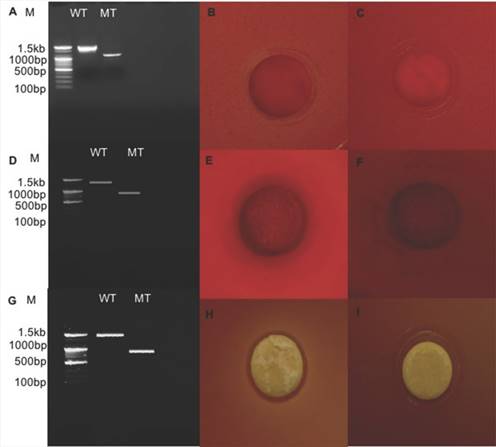
Relative expression level of all seven genes was confirmed by qRT-PCR (Fig. 2). In the first trial, cellulase, xylanase1 and esterase2 genes were mutagenized using CRISPR-cas9 system for 7 fibrinolytic enzyme genes. Each mutant was confirmed by PCR, which confirmed the target gene disturbed and reduced the size of amplicons. In addition, the mutant enzymatic activities were reduced and compared to the L. holzapfelli strain 5H4 in plate assays (Fig. 3). The fibrinolytic enzyme of L. holzapfelli strain 5H4 will be directly involved in fibrinolytic activity, indicating that L. holzapfelli strain 5H4 can improve NDF digestibility of the rye silage.
Nucleotide sequence accession number
L. holzapfelii strain 5H4 has been deposited in Korean Culture Center of Microorganisms under the number KCCM11788P and complete genome sequence was deposited at GenBank; accession number for the chromosome is SUB2178603.
Relative expression level of genes in L. holzapfelii 5H4. The housekeeping gene were pheS and rpoA.
Comparison of wild type and enzyme-related gene mutants (A) PCR analysis of cellulase region mutations using CRISPR-cas9 system. (B) cellulase enzyme assay in L. holzapfelii strain 5H4, (C) cellulase enzyme assay in celluase gene mutant, (D) PCR analysis of xylanase region, (E) xylanase enzyme assay in L. holzapfelii strain 5H4, (F) xylanase enzyme assay in xylanase gene mutant, (G) PCR assay of esterase region mutant, (H) esterase enzyme assay in L. holzapfelii strain 5H4, (I) esterase enzyme assay in esterase gene mutant.
Acknowledgements
This research was performed with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (PJ011012)” from the Rural Development Administration of Korea.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kim J, Chun J, Han HU. Leuconostoc kimchii sp. nov, a new species from kimchi. International Journal of Systematic Evolutionary Microbiology. 2000;50:1915-1919
2. Susiluoto T, Korkeala H, Bjorkroth KJ. Leuconostoc gasicomitatum is the dominating lactic acid bacterium in retail modified-atmosphere-packaged marinated broiler meat strips on sellby-day. International Journal of Systematic Evolutionary Microbiology. 2003;80:89-97
3. Leisner JJ, Vancanneyt M, Van der Meulen R. et al. Leuconostoc durionis sp. nov, a heterofermenter with no detectable gas production from glucose. International Journal of Systematic Evolutionary Microbiology. 2005;55:1267-1270
4. Antunes A, Rainey FA, Nobre MF. et al. Leuconostoc ficulneum sp nov, a novel lactic acid bacterium isolated from a ripe fig, and reclassification of Lactobacillus fructosus as Leuconostoc fructosum comb. nov. International Journal of Systematic Evolutionary Microbiology. 2002;52:647-655
5. Chambel L, Chelo IM, Ze-Ze L. et al. Leuconostoc pseudoficulneum sp nov, isolated from a ripe fig. International Journal of Systematic Evolutionary Microbiology. 2006;56:1375-1381
6. Bartolome B, Faulds CB, Tuohy M. et al. Influence of different xylanases on the activity of ferulic acid esterase of wheat bran. Biotechnology and Applied Biochemistry. 1995;22:65-73
7. Donaghy J, Kelly PF, McKay AM. Detection of ferulic acid esterase production by Bacillus spp. and Lactobacilli. Appllied Microbiology Biotechnology. 1998;50:257-260
8. Meslier V, Loux V, Renault P. Genome sequence of Leuconostoc pseudomesenteroides strain 4882, isolated from a dairy starter culture. Journal of Bacteriology. 2012;194:6637
Author contact
![]() Corresponding author: e-mail: kwakac.kr, tel: +82-55-772-1922, fax: +82-55-772-1929
Corresponding author: e-mail: kwakac.kr, tel: +82-55-772-1922, fax: +82-55-772-1929

 Global reach, higher impact
Global reach, higher impact