ISSN: 1839-9940
J Genomics 2014; 2:94-103. doi:10.7150/jgen.8043 This volume Cite
Review
How Do Y-Chromosomes Modulate Genome-Wide Epigenetic States: Genome Folding, Chromatin Sinks, and Gene Expression
Program in Molecular and Integrative Physiological Sciences, Department of Environmental Health, Harvard School of Public Health, Boston, MA 02115, USA.
Published 2014-5-1
Abstract
The Y chromosomes of Drosophila melanogaster and D. simulans contain only a handful of protein-coding genes, which are related to sperm mobility and reproductive fitness. Despite low or absent protein coding polymorphism, the Drosophila Y chromosome has been associated with natural phenotypic variation, including variation in the expression of hundreds to thousands of genes located on autosomes and on the X chromosome. Polymorphisms present in the large blocks of heterochromatin and consisting of differences in the amounts and kinds of sequences for satellite DNA and transposable elements may be the source of this modulation. Here we review the evidence and discuss mechanisms for global epigenetic regulation by repetitious elements in the Y chromosome. We also discuss how the discovery of this new function impacts the current knowledge about Y chromosome origin, its current dynamics, and future fate.
Keywords: Drosophila, epigenetics, gene regulation, Y-chromosome, YRV.
Introduction
The Y chromosome of D. melanogaster is a ~40 MB of segment of highly repetitive heterochromatic DNA that accounts for ~20% of the male haploid genome [1,2]. Early work identified six genetic loci (fertility factors) on the Y chromosome: two in the short arm (called ks-1 and ks-2) and four in the long arm (kl-1, kl-2, kl-3 and kl-5) [3,4]. Approaches combining bioinformatics and experimental analyses led to the molecular identification of several new Y-linked genes, some of which were correlated to the fertility factors [5-7]. A total of 13 protein-coding genes have been described so far: ARY, CCY (possible ks-2), kl-2 kl-3, kl-5, FDY, Mst77F (multiple copies), ORY (possible ks-1), Pp1-Y1, Pp1-Y2, Ppr-Y, PRY, WDY (possible kl-1) [5,6,8-11]. The genes have putative molecular function related to ATPase activity, motor activity, microtubule binding, protein serine/threonine phosphatase activity, and protein phosphatase type 1 regulator activity [12]. Putative biological functions for six of them (ARY, CCY, FDY, ORY, PRY and WDY) are more elusive. All Y-linked protein-coding genes are expressed exclusively in the testis and encode proteins that appear primarily related to sperm mobility and reproductive fitness [5,10-13].
Some Y-linked genes are megabase-sized; >100 times larger than the average size of autosomal or X-linked genes of D. melanogaster [1]. This is due to gigantic introns [14], whose molecular characterization remains to be determined. Overall, the chromosome is composed of large blocks of satellite DNA, transposable elements and, in D. melanogaster, the bobbed locus [1,15,16]. The bobbed locus consists of multiple tandem repeats (~30 to >240) coding for the ribosomal RNAs (rRNAs) [17]. Each rDNA unit consists of 3 rRNA subunits (18S, 5.8S and 28S), as well as transcribed (ITS1 and ITS2) and non-transcribed (IGS) spacers [17]. Another locus found in tens to hundreds of copies distributed in tandem on the Y chromosome of D. melanogaster is the Su(Ste), also called crystal [18]. Its basic unit consists of disrupted ORFs, an AT-rich region and the 1360 transposon [19]. Transcription of this gene produces small RNAs (piRNAs) that are necessary to inhibit the expression of the Ste gene located on the X chromosome [20,21].
Origin and evolution of the Y chromosome
There are two hypotheses to explain the origin of the Y chromosome of D. melanogaster. The canonical hypothesis explains the origin of sex chromosomes from one autosomal homologous pair [22-24]. The origin of the Y chromosome begins with the modification of a pair of autosomes to proto-sex chromosomes through the acquisition of sex determining loci [25-27]. The proto-Y might acquire a male sex-determining allele in one locus, and an allele that inhibits female's development in another locus. Lack of recombination between these loci is expected to be favored, such that suppression of recombination might spread through the chromosome. Unable to recombine with the X, the Y chromosome degenerates, losing many of its original genes, accumulating repetitive DNA and transposable elements and increasing its burden of heterochromatin.
The early stages of Y degeneration are especially well suited for understanding this process; neo-Y chromosomes that are formed by the fusion of an autosome to one of the sex chromosomes have been useful for analyses. Well-developed model systems include species in which the Y chromosome fused to an arm of an autosome, forming the so-called neo-Y [28]. This new chromosome is transmitted only to male offspring and is expected to degenerate with time. According to the canonical hypothesis, degeneration should be related to the age of these chromosome fusions. Newly formed neo-Y elements should have a large number of euchromatic functional genes and limited accumulation of mutations, while older neo-Ys are expected to contain few genes and many repetitive regions that result in larger amounts of heterochromatin. Three species of Drosophila have served as good candidates to test this hypothesis: D. albomicans, D. miranda and D. pseudoobscura. The species D. albomicans has a neo-Y newly formed, less than 120,000 years ago [29]. This neo-Y contains about 5,000 genes and hardly shows any signs of degradation [30,31]. The neo-Y of D. miranda originated about 1 million years ago, is partially heterochromatic, already shows accumulation of transposable elements [32,33], and has lost functionality in one third of its genes [34]. The species D. pseudoobscura has a neo-Y formed about 15 million years ago; it appears mostly heterochromatic and has 15 protein coding genes so far identified [35]. The temporal decay of neo-Ys fit with the expectation from the canonical model.
On the other hand, a non-canonical hypothesis states that the Y chromosome of D. melanogaster is not homologous to the X chromosome, but derived from a B chromosome [36] or the result of a partial duplication of an autosome [11]. The first evidence that the Y chromosome is not homologous to the X was reported by Brosseau (1960) [3], who noticed that X and Y chromosomes did not share genes in D. melanogaster. This observation is at odds with what would be expected if X and Y chromosomes had originated from the same autosomal pair. This investigator suggested that the Y chromosome could be novel; a specialized and unique element subjected to selection [3]. This view appears to have been shared by Lohe et al. (1993) [16] and Hackstein et al (1996) [36]. Lohe et al. (1993) [16] suggested that the Y chromosome could be more than a collection of degenerate genes, and functions of the chromosome could be due to recent amplification in repetitive DNA. Hackstein et al (1996) [36] explicitly argued that the Y chromosome could be a supernumerary B chromosome. Carvalho and colleagues have been recent proponents of the idea, with new studies and evidence accumulating in favor of such origin [11]. All protein-coding genes on the Y have paralogs on the autosomes and not on the X [5,6,8,9,11,37]. The same occurs for the Su(Ste) gene [38], which appears to be a rather recent acquisition to the Y-chromosome of D. melanogaster. All Y-linked protein-coding genes of D. melanogaster are distributed in autosomes in other Drosophila species [11]. Further evidence is the observation that the Y chromosome has interstitial telomeric regions, suggesting a merge with a supernumerary chromosome or translocation of telomeres [39]. Finally, in D. melanogaster and related species, the Y is not required for sex determination [40]. While the chromosome appears to acquire genes with male-related functions [6], these genic recruitments from autosomes seem to have occurred several times and on a gene-by-gene basis [6,37].
In any circumstance, it remains possible that the Y of D. melanogaster has lost the gene for sex determination and all homologous genes to the X chromosome. As pointed out by Kaiser and Bachtrog (2010) [41] and Carvalho et al. (2009) [11] resolution of the issue will require examination of sex determination systems across a larger set of Drosophila species and their ancestors. We envision that analyses of transitions to and from XO systems might shed important light towards addressing the dominant mode through which highly heterochromatic Y-chromosomes emerge in Drosophila.
Functions of the Y chromosome
XO males of D. melanogaster are viable but also sterile due to the inability to produce mature sperm in the morphologically normal testes [40,42]. However, engineered XXY female genotypes are for the most part undistinguishable from naturally occurring XX genotypes. Hence, it has been argued that the Y chromosome of D. melanogaster is an inert chromosome lacking functions besides its role in reproduction. Indeed, Y-linked protein-coding genes are only expressed in the male testis and required for sperm mobility and other spermatogenetic phenotypes [5,10-13]. Other Y-linked elements include the bobbed locus, which encodes ribosomal RNAs [17], and the crystal locus, which is responsible for the inhibition of the X-linked Ste gene that leads to male sterility when super expressed [20,21]. The rDNA locus appears to be responsible for X-Y pairing during meiosis [43,44].
Polymorphic phenotypes associated with the Y chromosome have also been documented. Genetic variation on the Y chromosome influences geotaxis [45], the regulation of gene expression in primary spermatocytes [46], the fitness of males [47], and the temperature sensitivity of spermatogenesis [48,49]. These results were surprising in part because population genetic theory and empirical data indicated that protein-coding genes in the haploid Y chromosome were minimally variable relative to the rest of the genome [50,51]; the expectation of meager variation arises in part due to the absence of recombination as well as lower population sizes of Y chromosomes. Partial sequencing of protein-coding genes, including their introns, in populations of D. melanogaster showed that these genes have indeed very low nucleotide diversity [52,53]. Chippindale and Rice (2001) [47] suggested that variation in total fitness of males could be due to polymorphism in protein-coding genes, which might have remained undetected due to the small region sequenced by Zurovcova and Eanes (1999) [52]. A contribution from protein-coding variation might indeed be non-negligible, as models with high epistasis are likely to raise the expectation for Y-linked variation in segments of typical mutation rate such as protein coding genes. Rohmer et al. (2004) [48] reasoned that the explanation for the role of polymorphic Y-linked variation in the sensitivity of spermatogenesis to temperature could emerge from a gene with yet unknown function. This is also a likely possibility.
In any circumstance, the low number of Y-linked genes and their low genetic diversity remained hard to reconcile with data suggesting polymorphic phenotypic variation associated with the Y chromosome. One clue came from investigations into the epigenetic regulatory component of the D. melanogaster Y chromosome. This is because Y chromosome aneuploidies have long been known to cause suppression of position-effect variegation (PEV) [54]. One form of PEV occurs when euchromatic genes are moved from their native location to a new site near a heterochromatic segment [55].
These observations point to intriguing mechanisms by which heterochromatic Y-chromosomes influence phenotypes and PEV. For instance, groundbreaking studies by Zhang et al. (2000) [46] identified the involvement of repetitive sequences on the long arm of the Y chromosome in the regulation of gene expression in primary spermatocytes. More recently, it has been suggested that Y chromosome polymorphisms could be associated with phenotypic variation in the absence of nucleotide variation for protein-coding segments [56-59]. The evidence emerged from studies that examined variation in gene expression between D. melanogaster strains that differed only in the origin of the Y chromosome [56]. The phenomenon of quantitative effects of polymorphic Y-chromosomes in the expression of hundreds to thousands of genes located on autosomes and on the X chromosome is called Y-linked regulatory variation (YRV). These data showed that the Y chromosome is genetically variable and responsible for regulating genes in all chromosomes. So where was the variation? One possibility is that the causative variation might be the large blocks of heterochromatin [56,57], composed of sequences of repetitive DNA and transposable elements, which constitute the majority of the Y chromosome.
If the regulatory activity of the Y chromosome is actually mediated by non-protein coding heterochromatic elements rather than the product of protein-coding genes, such activity might occur in XXY females and in somatic cells of XY males [58]. This is because protein-coding genes are expressed only in the germ cells of males. In agreement with the hypothesis of a non-genic basis for YRV, the phenomenon is strongly manifested in XXY females and in male somatic tissues [59]. The Y-chromosome is heterogeneous and composed of diverse sequence segments. This raises the question of which specific segment might be causative of YRV. Transposable elements may interfere in the regulation of gene expression [60,61], and variable transposable element activity from within the Y-chromosome might be one component of YRV. Another candidate is the bobbed locus. Deletions of rDNA units from the D. melanogaster Y chromosome affect the expression of autosomal and X-linked genes [62-64]. However, the rDNA locus can at best explain ~20-40% of YRV in D. melanogaster [63], and cannot explain YRV in D. simulans [65], because this latter species lacks Y-linked rDNA loci. Hence, the data suggest the involvement of multiple loci contributing to the emergence of YRV [59].
Several studies have further described the characteristics of YRV. First, it is rapidly evolving. After 550 generations, lineages derived from a single Y already presented abundant YRV [64]. Furthermore, the role of the Y chromosome in suppressing PEV suggests that it is important for modulating chromatin states in the genome [59]. Second, classical chromatin components interact with YRV. The heterochromatin protein 1 (HP1a) is a crucial component of chromatin and the protein Mod(mdg4) is a classic chromatin modifying factor. Both of these are necessary for the manifestation of testicular YRV in D. melanogaster [66]. Altogether, the genes affected by the Y chromosome are, in general, expressed tissue specifically, their expression diverges rapidly, they are biased towards testis expression, they are clustered in coherent gene ontology categories, they are not physically linked, and their expression is dependent on the genomic background of autosomes [56,59,63,64,66-68].
Finally, the Y chromosome also affects gene expression through mechanisms that are reminiscent of genomic imprinting [69]. The imprinting of the Y modulates the epigenetic process of dosage compensation and paternal imprinting increases the regulatory ability of the Y [70]. Males of D. melanogaster that differ only in the maternal or paternal origin of X and Y chromosomes exhibit differential expression in hundreds of genes mainly in testis and midgut cells [68]. These studies suggest that the Y-chromosome is differentially modified in the epigenetic context of the male testis and female ovary, with widespread consequence to the rest of the genome.
Molecular mechanisms for Y-linked regulatory variation
A variety of mechanistic hypotheses can be conceived to explain YRV's mode of action. Two of them involve transcription factors encoded by Y-linked genes [58]. These transcription factors could bind directly to target genes, or could interact with other proteins that bind to target genes. These interactions facilitate or prevent the binding of RNA-polymerase II to DNA and thus, expression of target genes would be facilitated or inhibited (Figure 1). Another hypothesis is related to intron size variation [58]. Since some Y-linked genes have gigantic introns, it is conceivable that variation in intron size could lead to differential transcription of these introns and titration of transcription cofactors. Transcription of Y-linked protein-coding genes occurs only in spermatogenesis [11] and thus it is assumed that their products should preferentially influence male traits. These hypotheses are supported by the fact that YRV affects genes that are highly expressed in males and less expressed in females [56]. However, YRV is also manifested in XXY females, and in somatic tissues of males [59], which do not express Y-linked genes [73]. Moreover, as already mentioned, low polymorphism was detected in the coding region of the Y [52,53]. Hence, a significant component of YRV is bound to be independent of the expression of Y linked protein coding genes [59]. Nevertheless, elucidating the role of Y-linked protein-coding genes would be crucial: genes of unknown function might contribute to transcription factor activity and might harbor low levels of coding sequence polymorphism or expression variation. If a component of YRV is caused by transcription factors encoded by Y-linked genes then these genes must also be present in other species showing YRV such as D. simulans (see below). From the six genes with lesser-known function in D. melanogaster, five are also found on the Y chromosome of D. simulans [11,37].
The classical hypothesis, known as the heterochromatin sink model [74,75], suggests that the content or the length of heterochromatin blocks can serve as a sink for transcription factors or chromatin regulators, resulting in their depletion or redistribution throughout the rest of the genome. The model might underlie YRV if the copy number of repetitious elements is variable [56] and those elements serve as a binding site for proteins that modulate the switch between euchromatic and heterochromatic states in PEV [55]. The Y chromosome might compete for and recruit proteins that target heterochromatin, and consequently hinder the spread of heterochromatin elsewhere [76]. Available evidence with lacZ reporter gene indicates that genetic elements located in a specific segment of the long arm of the Y chromosome might have the ability to modulate gene expression [46]. The identified segment is relatively large and the causative factors appear dispersed within it. Intriguingly, (AAGAG)n and (AAAGAGA)n repeats located on the Y are binding sites for the transcription factor GAGA, and raise the interesting possibility that the regulation might be mediated through GAGA-factor [46]. Another example refers to the transcription factor D1. It binds preferentially to satellite DNA, and variation on the Y chromosome could lead to the modulation of its availability elsewhere (Figure 2). Another possibility involves HP1a, a protein whose cascading effects on gene regulation are not only limited to models of competition through chromatin sinks. HP1a is a core component of heterochromatin, and it regulates the expression of hundreds of genes [76]. HP1a can also directly bind to RNA molecules, and interact with RNA-polymerase II as well as with heterogeneous nuclear ribonucleoproteins involved in RNA processing [77]. It is possible that the differential deposition of heterochromatin due to Y polymorphism can influence the availability of HP1a and lesser binding on its mRNA targets. Addressing if HP1a, for example, might be differentially bound to specific positions in flies differing only in their Y might be a necessary step to address the mechanics of how YRV is manifested. Finally, the Y chromosome could serve as a scaffold for protein complexes that interact with splicing factors influencing the processing of mRNA of the target genes [71,72].
YRV molecular mechanisms: models based on regularory factors encoded by Y-linked genes. Y chromosomes are colored and autosomes are in black. Only one copy of each chromosome is represented. √ means activation and û means inhibition of gene expression. A: Regulatory factors (orange squares) from the Y chromosome (blue) facilitate or prevent the expression of target genes. B: A different Y chromosome (red) encodes a different regulatory factor (orange circles) or express the same set of factors at different dose.
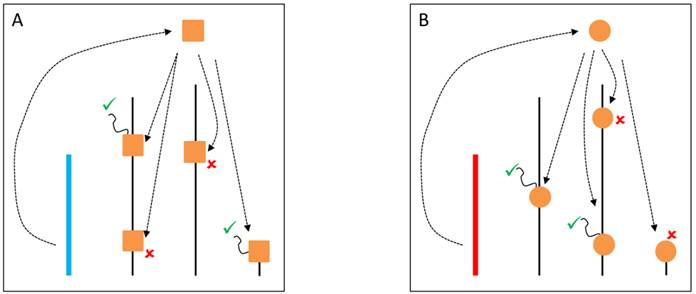
YRV molecular mechanisms: models based on the heterochromatin sink. Y chromosomes are colored and autosomes are in black. Only one copy of each chromosome is represented. √ means activation and û means inhibition of gene expression. A: Regulatory factors (triangles) bind on satellite DNA in the Y chromosome (blue) and are depleted at other sites of the genome. The redistribution causes global modulation of gene expression. B: Regulatory factors might bind in other positions and with different strenghts in another Y chromosome (red) leading to a differential pattern of depletion across the genome.
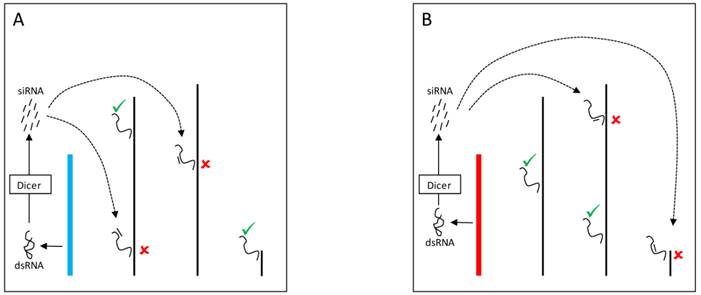
The mechanisms by which the Y chromosome produces genome-wide gene expression modulation might also be related to the expression of small RNAs [56]. Indeed, multiple pathways might mediate these effects and can account for the modulation of PEV as well as for direct effects on gene expression. In one pathway, double stranded RNA (dsRNA) transcribed from elements in the Y chromosome might be cleaved by the Dicer enzyme into siRNAs (Figure 3). The dsRNA is converted into siRNAs that assembles into the RNA-induced initiation of transcriptional gene silencing complex (RITS) [78,79]. This complex can target specific segments of the genome and draw the protein SU(VAR)3-9 methyltransferase that modifies the lysine 9 of histone H3 (H3K9). The modification creates a binding site for the protein HP1a, which in turn attracts other proteins required for heterochromatin formation [76,78].
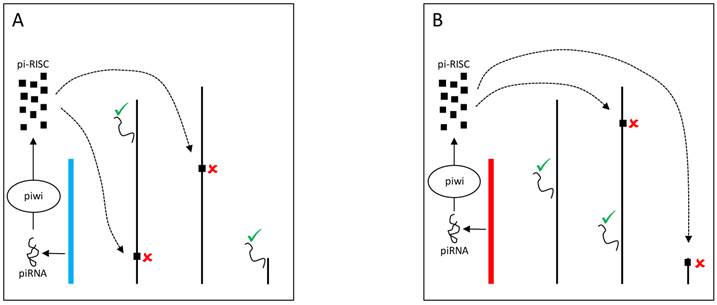
The Y chromosome can also be a source of piRNAs and modulate gene expression through Dicer-independent mechanisms. Piwi proteins assemble with small RNAs (piRNAs) into the piRNA-induced silencing complex (pi-RISC) [80] in the nucleus. The complex presumably binds to transposable elements in the other chromosomes and mediates the recruitment of heterochromatinizing proteins that silence transposable elements targets and their neighboring genes (Figure 4) [81]. Evidence suggests that YRV might disproportionately modulate families of transposable elements [56]. Variation in the satellite DNAs or transposable elements might underlie variable piRNA populations and consequently lead to the silencing of different genes targets. Establishing if small RNAs are involved in YRV might require detection of RNAs derived from transposable elements and other repetitive DNA from the Y chromosome. The behavior of the Y chromosome heterochromatin might be similar to elements in centromeric region of autosomes, but the characterization of repetitive elements has remained a challenge.
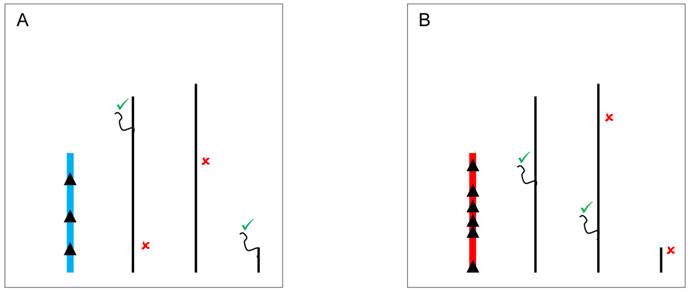
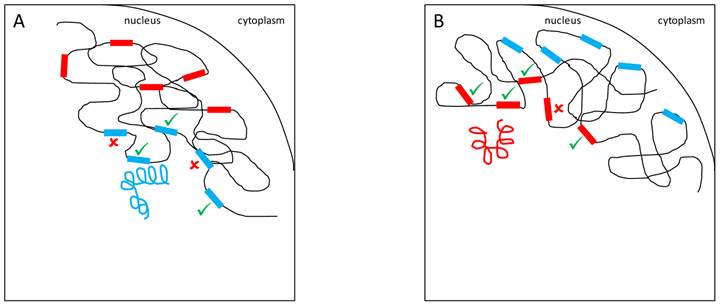
Another hypothesis, the spatial arrangement model, states that Y-linked variation might alter chromosomal territories and packaging in the nucleus. Indeed, it is well-established that the position of genes and chromosomes influences gene expression [58,82,83]. For example, human Hox genes are inactive when on the periphery of the nucleus but are repositioned to the center of the nucleus for transcription to occur [84]. This movement probably occurs towards locations where molecules responsible for gene expression are concentrated, the transcription centers [85]. Hence, variable Y chromosomes might exert their effects through changes in the spatial positioning of X-linked and autosomal genes (Figure 5).
YRV molecular mechanisms: models based on RNAi. Y chromosomes are colored and autosomes are in black. Only one copy of each chromosome is represented. √ means activation and û means inhibition of gene expression. A: Double stranded RNA (dsRNA) transcribed from transposable elements and/or pseudogenes of the Y chromosome (blue) are postulated to be cleaved by the Dicer enzyme into siRNAs. These siRNAs might bind to target mRNAs which would be degraded by the RNA-induced silencing complex. B: Variation in the satellite DNAs or transposable elements from a different Y chromosome (red) might lead to different dsRNA populations, different siRNAs, and consequently the silencing of different target genes.
YRV molecular mechanisms: models based on piRNA. Y chromosomes are colored and autosomes are in black. Only one copy of each chromosome is represented. √ means activation and û means inhibition of gene expression. A: piRNAs from the Y chromosome (blue) interact with Piwi proteins in the nucleus assembling the piRNA-induced silencing complex (pi-RISC). This complex binds to transposable elements in the other chromosomes and leads to the recruitment of heterochromatinizing proteins, silencing the transposable elements targets and their neighbouring genes. B: Variation in the piRNAs source (Y chromosome in red) would lead to different pi-RISC population and consequently the silencing of different target genes.
YRV molecular mechanisms: Models based on spatial arrangement. Y chromosomes are colored and autosomes are in black. √ means activation and û means inhibition of expression. A: Y chromosome packaging (blue spiral) in the nucleus and its target genes (blue rectangles) in other chromosomes. B: A different Y chromosome (red spiral) might alter chromatin packaging and the positioning of target genes (red rectangles) in the nuclear environment.
YRV in other Drosophila species and in mammals
Phenotypic diversity caused by Y-linked variation is not restricted to D. melanogaster. In D. virilis, the Y chromosome has effects on the variation of courtship song characters [86]. In D. simulans, the Y chromosome modulates the ability to suppress the sex-ratio distortion induced by the X chromosome [57,65], and also exerts regulatory consequences [65]. These results also raise the question of whether non-Drosophila organisms that have a heterochromatin-rich Y chromosome with few genes would also display YRV [56]. In mice, the Y has no influence on PEV and does not appear to be a sink for proteins with high affinity for heterochromatin [87]; in humans, substantial variation in heterochromatin amount between different Y-chromosome haplogroups has been reported [88]. While evidence suggestive of YRV was found in the euchromatic parts of the human Y chromosome and in experiments in mice models, the genic and non-genic components of YRV are harder to disentangle in these systems [87,89]. The expression or the copy number of protein-coding genes on the Y is likely to have a regulatory role in the expression of autosomal genes, by alternative splicing or modification of the chromatin, for instance [87,89]. In any circumstance, non-genic components of YRV as suggested in Drosophila might also be relevant for heterochromatic segments in the human Y chromosome. Finally, the larger inactive X chromosome, which is heterochromatized in females, might be a more effective player exerting heterochromatin sink pressure in mammals.
Final considerations
The Y chromosome of D. melanogaster is responsible for male fertility and is not invariant within populations. Instead, the chromosome is composed of polymorphic regions that play regulatory roles. Y-linked regulatory variation (YRV) refers to the global modulation of gene expression due to Y chromosome polymorphism and divergence. YRV is also detectable in mammals but it remains unclear if the mammalian Y chromosome exerts its regulatory consequences through mechanisms similar to those exerted by the Drosophila Y chromosome. Evidence in Drosophila suggests a non-genic component to the phenomenon. There is a potential difference between the YRV of humans and Drosophila: the regulatory role of heterochromatin, which might have a lesser role in human YRV, appears to be essential for YRV in Drosophila. In any circumstance, molecular mechanisms of YRV are poorly elucidated in all organisms; we are tempted to speculate that YRV in mammals might be manifested through the Y-linked modulation of X-linked heterochromatin states in female progeny.
Evolutionary questions regarding YRV are also evident. When did the Y chromosome start to perform its regulatory function? The neo-Y of D. albomicans, D. miranda and D. pseudoobscura, which were formed at different times and with different amounts of heterochromatin [29,31,32,35], have provided data that match the expected narrative for Y chromosome degeneration. Extension to other systems might be interesting to shed light on YRV. For instance, it would be interesting to ascertain if the Y chromosomes that arose independently in two species of fish of the genus Oryzias [90] have evolved regulatory roles. Also, studies documenting variation within and between D. melanogaster and D. simulans populations could be informative.
Regardless of how the Y chromosome originated, it has evolved to become a source of gene expression variation in D. melanogaster males, and its current structure shares several characteristics with B chromosomes. Carvalho et al. (2009) [11] argued that the Y chromosome in D. melanogaster is a product of duplications of autosomal genes. B chromosomes might also originate this way [91,92] and it appears entirely possible that the Y of D. melanogaster originated from a B chromosome. Sex chromosomes (Y or W) that could have originated from B chromosomes were described for the frog Leiopelma hochstetteri [93] and the insect Cacopsylla peregrina [94]. In D. albomicans, B chromosomes might confer adaptive advantage; individuals carrying 1 or 2 B chromosomes produce more offspring than those without them [30]. Intriguingly, it has been observed that heterochromatic regions from B chromosomes of rye (Secale cereale) have the ability to regulate the expression of non-B genes [95]. In addition, an explanation of how the Bs of rye would perform this regulation might involve modulation of the spatial organization of the genome [95].
Finally, we expect that selection on the regulatory functions of the Y chromosome might not be negligible, and that degeneration might be a misnomer to describe the attributes of the chromosome. Y-linked transposable elements and repetitive DNA might not only be consequences of relaxation of selection on the Y chromosome, but they might also be positively selected through their effects on genome-wide epigenetic regulation. Evidence that transposable elements participate in the formation of heterochromatin is abundant e.g. [32,96,97] and it is straighforward to imagine how positive and negative natural selection might contribute to emegence of functional variation in this system. Moreover, transposable elements distribution on the Y chromosome of D. melanogaster might not be random, and insertion location could contribute to some Y chromosome phenotypes [98], including trans-regulatory epigenetic variation. In conclusion, the Y chromosome and its cryptic variation is expected to be important sources of adaptive and non-adaptive phenotypic variation in Drosophila as well as mammals. Altogether, the regulatory role of the Y chromosome is bound to be crucial in shaping how this chromosome has evolved, its current phenotypic consequences, and its long-term fate.
Acknowledgements
We thank A. Bernardo Carvalho for comments on the manuscript and members of the Lemos lab for discussions. BL thanks support from the Ellison Medical Foundation New Scholar in Aging Award and the Smith Family Award for Biomedical Research.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gatti M, Pimpinelli S. Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster. Chromosoma. 1983;88:349-73
2. Hoskins RA, Smith CD, Carlson JW, Carvalho AB, Halpern A, Kaminker JS, Kennedy C, Mungall CJ, Sullivan BA, Sutton GG, Yasuhara JC, Wakimoto BT, Myers EW, Celniker SE, Rubin GM, Karpen GH. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002;3:research0085.1-0085.16
3. Brosseau GE. Genetic analysis of the male fertility factors on the Y chromosome of Drosophila melanogaster. Genetics. 1960;45:257-74
4. Kennison JA. The genetic and cytological organization of the Y chromosome of Drosophila melanogaster. Genetics. 1981;98:529-48
5. Carvalho AB, Lazzaro BP, Clark AG. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci U S A. 2000;97:13239-44
6. Carvalho AB, Dobo BA, Vibranovski MD, Clark AG. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:13225-30
7. Carvalho AB, Vibranovski MD, Carlson JW, Celniker SE, Hoskins RA, Rubin GM, Sutton GG, Adams MD, Myers EW, Clark AG. Y chromosome and other heterochromatic sequences of the Drosophila melanogaster genome: how far can we go? Genetica. 2003;117:227-37
8. Vibranovski MD, Koerich LB, Carvalho AB. Two new Y-linked genes in Drosophila melanogaster. Genetics. 2008;179:2325-7
9. Krsticevic FJ, Santos HL, Januário S, Schrago CG, Carvalho AB. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics. 2010;184:295-307
10. Gepner J, Hays TS. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci U S A. 1993;90:11132-6
11. Carvalho AB, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009;25:270-7
12. Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond JR, Strelets VB, Wilson RJ, Consortium F. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751-D757
13. Goldstein LS, Hardy RW, Lindsley DL. Structural genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982;79:7405-9
14. Bonaccorsi S, Pisano C, Puoti F, Gatti M, Moro PA. Y chromosome loops in Drosophila melanogaster. Genetics. 1988;120:1015-34
15. Bonaccorsi S, Lohe A. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: relationships between satellite sequences and fertility factors. Genetics. 1991;129:177-89
16. Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149-74
17. Lyckegaard EMS, Clark AG. Evolution of ribosomal RNA gene copy number on the sex chromosomes of Drosophila melanogaster. Mol Biol Evol. 1991;8:458-74
18. Livak KJ. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics. 1990;124:303-16
19. Balakireva MD, Shevelyov YY, Nurminsky DI, Livak KJ, Gvozdev VA. Structural organization and diversification of Y-linked sequences comprising Su(Ste) genes in Drosophila melanogaster. Nucleic Acids Res. 1992;20:3731-6
20. Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA. 2010;16:2503-15
21. Kalmykova AI, Dobritsa AA, Gvozdev VA. Su(Ste) diverged tandem repeats in a Y chromosome of Drosophila melanogaster are transcribed and variously processed. Genetics. 1998;148:243-9
22. Muller HJ. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics. 1918;3:422-99
23. Muller HJ. Further studies on the nature and causes of gene mutations. Proc 6th Int Congr Genet. Ithaca, USA. 1932:213-55
24. Ohno S. Sex chromosomes and sex-linked genes. Berlin, Germany: Springer. 1967
25. Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030-3
26. Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118-28
27. Charlesworth D. Plant sex chromosome evolution. J Exp Bot. 2013;64:405-20
28. Kaiser VB, Zhou Q, Bachtrog D. Nonrandom gene loss from the Drosophila miranda neo-Y chromosome. Genome Biol Evol. 2011;3:1329-37
29. Bachtrog D. The speciation history of the Drosophila nasuta complex. Genet Res. 2006;88:13-26
30. Zhou Q, Zhu H, Huang Q, Zhao L, Zhang G, Roy SW, Vicoso B, Xuan Z, Ruan J, Zhang Y, Zhao R, Ye C, Zhang X, Wang J, Wang W, Bachtrog D. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics. 2012;13:109
31. Cheng C-H, Chang C-H, Chang H. Early-stage evolution of the neo-Y chromosome in Drosophila albomicans. Zool Stud. 2011;50:338-49
32. Steinemann M, Steinemann S. Enigma of Y chromosome degeneration: Neo-Y and Neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica. 1998;102-103:409-20
33. Steinemann S, Steinemann M. Biased distribution of repetitive elements: a landmark for neo-Y chromosome evolution in Drosophila miranda. Cytogenet Cell Genet. 2001;93:228-33
34. Bachtrog D. Sex chromosome evolution: molecular aspects of Y-chromosome degeneration in Drosophila. Genome Res. 2005;15:1393-401
35. Carvalho AB, Clark AG. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science. 2005;307:108-10
36. Hackstein JHP, Hochstenbach R, Hauschteck-Jungen E, Beukeboom LW. Is the Y chromosome of Drosophila an evolved supernumerary chromosome? BioEssays. 1996;18:317-23
37. Koerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949-51
38. Kalmykova AI, Shevelyov YY, Dobritsa AA, Gvozdev VG. Acquisition and amplification of a testis-expressed autosomal gene, SSL, by the Drosophila Y chromosome. Proc Natl Acad Sci U S A. 1997;94:6297-302
39. Abad JP, de Pablos B, Agudo M, Molina I, Giovinazzo G, Martín-Gallardo A, Villasante A. Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere-derived sequences at internal regions. Chromosoma. 2004;113:295-304
40. Bridges CB. Non-disjunction as proof of the chromosome Theory of Heredity. Genetics. 1916;1:1-52
41. Kaiser VB, Bachtrog D. Evolution of sex chromosomes in insects. Annu Rev Genet. 2010;44:91-112
42. Muller HJ. A gene for the fourth chromosome of Drosophila. J Exp Zool. 1914;17:325-36
43. McKee BD, Karpen GH. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell. 1990;61:61-72
44. McKee BD, Satter MT. Structure of the Y chromosomal Su(Ste) locus in Drosophila melanogaster and evidence for localized recombination among repeats. Genetics. 1996;142:149-61
45. Stoltenberg SF, Hirsch J. Y-chromosome effects on Drosophila geotaxis interact with genetic or cytoplasmic background. Anim Behav. 1997;53:853-64
46. Zhang P, Timakov B, Stankiewicz RL, Turgut IY. A trans-activator on the Drosophila Y chromosome regulates gene expression in the male germ line. Genetica. 2000;109:141-50
47. Chippindale AK, Rice WR. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2001;98:5677-82
48. Rohmer C, David JR, Moreteau B, Joly D. Heat induced male sterility in Drosophila melanogaster: adaptive genetic variations among geographic populations and role of the Y chromosome. J Exp Biol. 2004;207:2735-43
49. David JR, Araripe LO, Chakir M, Legout H, Lemos B, Pétavy G, Rohmer C, Joly D, Moreteau B. Male sterility at extreme temperatures: a significant but neglected phenomenon for understanding Drosophila climatic adaptations. J Evol Biol. 2005;18:838-46
50. Clark AG. Variation in Y chromosome segregation in natural populations of Drosophila melanogaster. Genetics. 1987;115:143-51
51. Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc B. 2000;355:1563-72
52. Zurovcova M, Eanes WF. Lack of nucleotide polymorphism in the Y-linked sperm flagellar dynein gene Dhc-Yh3 of Drosophila melanogaster and D. simulans. Genetics. 1999;153:1709-15
53. Larracuente AM, Clark AG. Surprising differences in the variability of Y chromosomes in African and cosmopolitan populations of Drosophila melanogaster. Genetics. 2013;193:201-14
54. Gowen JW, Gay EH. Chromosome constitution and behavior in eversporting and mottling in Drosophila melanogaster. Genetics. 1934;19:189-208
55. Dimitri P, Pisano C. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the ammount of Y chromosome. Genetics. 1989;122:793-800
56. Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91-3
57. Montchamp-Moreau C, Ginhoux V, Atlan A. The Y chromosomes of Drosophila simulans are highly polymorphic for their ability to suppress sex-ratio drive. Evolution. 2001;55:728-37
58. Malone JH, Oliver B. The sex chromosome that refused to die. BioEssays. 2008;30:409-11
59. Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci U S A. 2010;107:15826-31
60. Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33:102-6
61. Puig M, Cáceres M, Ruiz A. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc Natl Acad Sci U S A. 2004;101:9013-8
62. Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci U S A. 2009;106:17829-34
63. Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7:e1001376
64. Zhou J, Sackton TB, Martinsen L, Lemos B, Eickbush TH, Hartl DL. Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila. Proc Natl Acad Sci U S A. 2012;109:9941-6
65. Branco AT, Tao Y, Hartl DL, Lemos B. Natural variation of the Y chromosome suppresses sex ratio distortion and modulates testis-specific gene expression in Drosophila simulans. Heredity. 2013;111:8-15
66. Branco AT, Hartl DL, Lemos B. Chromatin-associated proteins HP1 and Mod(mdg4) modify Y-linked regulatory variation in the Drosophila testis. Genetics. 2013;194:609-18
67. Jiang P-P, Hartl DL, Lemos B. Y not a dead end: epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics. 2010;186:109-18
68. Lemos B, Branco AT, Jiang P-P, Hartl DL, Meiklejohn CD. Genome-wide gene expression consequences of sex chromosome imprinting in Drosophila. G3 (Bethesda). 2014;4(1):1-10
69. Maggert KA, Golic KG. The Y chromosome of Drosophila melanogaster exhibits chromosome-wide imprinting. Genetics. 2002;162:1245-58
70. Menon DU, Meller VH. Imprinting of the Y chromosome influences dosage compensation in roX1 roX2 Drosophila melanogaster. Genetics. 2009;183:811-20
71. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925-38
72. Venkatesh S, Workman JL, Smolle M. UpSETing chromatin during non-coding RNA production. Epigenet Chromat. 2013;6:16
73. Ashburner M, Golic KG, Hawley RS. Drosophila: a laboratory handbook, 2nd ed. Cold Spring Harbor, USA: Cold Spring Harbor Laboratory Press. 2005
74. Zuckerkandl E. A possible role of “inert” heterochromatin in cell differentiation. Action of and competition for “locking” molecules. Biochimie. 1974;56:937-54
75. Henikoff S. Dosage-dependent modification of position-effect variegation in Drosophila. BioEssays. 1996;18:401-9
76. Schulze SR, Wallrath LL. Gene regulation by chromatin structure: paradigms established in Drosophila melanogaster. Annu Rev Entomol. 2007;52:171-92
77. Piacentini L, Fanti L, Negri R, Del Vescovo V, Fatica A, Altieri F, Pimpinelli S. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5:e1000670
78. Elgin SCR, Grewal SIS. Heterochromatin: silence is golden. Curr Biol. 2003;13:R895-R898
79. Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897-1901
80. Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656-68
81. Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964-80
82. Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 1996;381:529-31
83. Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787-800
84. Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119-30
85. Cook PR. The organization of replication and transcription. Science. 1999;284:1790-5
86. Huttunen S, Aspi J. Complex inheritance of male courtship song characters in Drosophila virilis. Behav Genet. 2003;33:17-24
87. Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132-40
88. Navarro-Costa P, Plancha CE. Heterochromatin: the hidden epigenetic geography of the Y chromosome. Hum Reprod Update. 2011;17:434
89. Case LK, Wall EH, Dragon JA, Saligrama N, Krementsov DN, Moussawi M, Zachary J, Huber SA, Blankenhorn EP, Teuscher C. The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res. 2013;23(9):1474-85
90. Takehana Y, Demiyah D, Naruse K, Hamaguchi S, Sakaizumi M. Evolution of different Y chromosomes in two medaka species, Oryzias dancena and O. latipes. Genetics. 2007;175:1335-40
91. Martis MM, Klemme S, Banaei-Moghaddam AM, Blattner FR, Macas J, Schmutzer T, Scholz U, Gundlach H, Wicker T, Šimková H, Novák P, Neumann P, Kubaláková M, Bauer E, Haseneyer G, Fuchs J, Doležel J, Stein N, Mayer KFX, Houben A. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc Natl Acad Sci U S A. 2012;109:13343-6
92. Yoshida K, Terai Y, Mizoiri S, Aibara M, Nishihara H, Watanabe M, Kuroiwa A, Hirai H, Hirai Y, Matsuda Y, Okada N. B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 2011;7:e1002203
93. Green DM. Cytogenetics of the endemic New Zealand frog, Leiopelma hochstetteri: extraordinary supernumerary chromosome variation and a unique sex-chromosome system. Chromosoma. 1988;97:55-70
94. Nokkala S, Grozeva S, Kuznetsova V, Maryanska-Nadachowska A. The origin of the achiasmatic XY sex chromosome system in Cacopsylla peregrina (Frst.) (Psylloidea, Homoptera). Genetica. 2003;119:327-32
95. Banaei-Moghaddam AM, Meier K, Karimi-Ashtiyani R, Houben A. Formation and expression of pseudogenes on the B chromosome of rye. Plant Cell. 2013;25:2536-44
96. Danilevskaya O, Lofsky A, Kurenova EV, Pardue ML. The Y chromosome of Drosophila melanogaster contains a distinctive subclass of Het-A-related repeats. Genetics. 1993;134:531-43
97. Steinemann S, Steinemann M. Y chromosomes: born to be destroyed. BioEssays. 2005;27:1076-83
98. Gvozdev VA, Kogan GL, Usakin LA. The Y chromosome as a target for acquired and amplified genetic material in evolution. BioEssays. 2005;27:1256-562
Author contact
![]() Corresponding author: Bernardo Lemos, 665 Huntington Avenue, Bldg 2, Rm 219, Harvard School of Public Health, Boston, MA 02115-6021. Email: blemosharvard.edu.
Corresponding author: Bernardo Lemos, 665 Huntington Avenue, Bldg 2, Rm 219, Harvard School of Public Health, Boston, MA 02115-6021. Email: blemosharvard.edu.

 Global reach, higher impact
Global reach, higher impact