ISSN: 1839-9940
J Genomics 2014; 2:31-44. doi:10.7150/jgen.5260 This volume Cite
Review
Molecular Heterogeneities of Adipose Depots - Potential Effects on Adipose-Muscle Cross-Talk in Humans, Mice and Farm Animals
1. Research Unit Muscle Biology and Growth, Leibniz-Institute for Farm Animal Biology (FBN), W.-Stahl-Allee 2, D-18196 Dummerstorf, Germany;
2. Research Unit Molecular Biology, Leibniz-Institute for Farm Animal Biology (FBN), W.-Stahl-Allee 2, D-18196 Dummerstorf, Germany;
3. Department of Animal Sciences, Washington State University, Pullman, WA 99164, USA.
Published 2014-1-20
Abstract
Adipose tissue is considered as a major endocrine organ that secretes numerous proteins called adipokines. The heterogeneous nature of adipose tissue in different parts of the body suggests respective heterogeneity of proteomes and secretomes. This review consolidates knowledge from recent studies targeting the diversity of different adipose depots affecting the pattern of secreted adipokines and discusses potential consequences for the cross-talk between adipose and skeletal muscle in humans, rodent models and farm animals. Special attention is paid to muscle-associated fat depots like inter- and intramuscular fat that become focus of attention in the context of the rather new notion of skeletal muscle as a major endocrine organ. Understanding the complexity of communication between adipocytes and skeletal muscle cells will allow developing strategies for improvement of human health and for sustainable production of high quality meat.
Keywords: adipose depots, intramuscular fat, adipokines, myokines, review.
Introduction
Obesity is considered as an expanding epidemic in many developed countries and it is associated with major adverse consequences on human health. By targeting this problem, research has dramatically advanced our knowledge on the regulation of energy balance and physiology of adipose tissue during the last decades [1]. For a long time, adipose tissue was perceived as a merely passive lipid storage depot that buffers a fluctuating energy supply. A new concept emerged when it was discovered that adipocytes produce adipsin and secrete leptin (reviewed in [1-3]). By now, adipose tissue is considered a major endocrine organ, with more than 600 potentially secreted proteins identified so far [2, 4, 5]. These proteins, termed adipokines, exert endo-, para-, and autocrine effects and have been linked to biological processes like metabolism, immunity and fertility [6-8]. Furthermore, relationships with adverse health conditions like metabolic syndrome, cardiovascular disease, psoriasis and dementia have been established [9-12].
In humans and rodents, the two major fat depots include visceral adipose tissue (VAT), which is the fat of the intra-abdominal cavity, and subcutaneous adipose tissue (SAT), which is the fat layer under the skin [13]. In addition to VAT and SAT, intermuscular adipose tissue (IMAT) and intramuscular fat (IMF) are also measured in farm animal species such as swine and cattle. The former is the fat located between distinct muscles, while the latter is the adipose tissue within a muscle [14]. The terminology for fat depots and the methods for their analysis are summarized in Table 1. The different use of terms is mainly due to applied analysis techniques and, to a certain degree, to different muscle groups considered in humans and animals. Indirect measurements of fat depots with imaging and spectroscopy technologies prevail in investigations in human. In meat animals and rodents, different depots are readily accessible for analysis post-mortem. Muscle related fat is analyzed mainly in human limbs, whereas fat in large dorsal muscles is the focus of animal science. Traditional selection of farm animals aimed at reducing body fat content and increasing muscle mass over the long term. Besides the desired reduction of adipose tissue not suited for human consumption, the correlated decrease in the fat content in skeletal muscle resulted in reduced palatability of meat [23-25]. This in turn, caused animal scientists to elucidate adipogenic mechanisms in farm animals for manipulation of IMF (reviewed in [26]). Composition as well as distribution and amount of IMF have been the targets because these parameters equally influence the eating quality of meat [15].
Although research in human and farm animals ultimately aims at different goals - identification of potential drug targets for metabolic diseases versus optimization of meat quality - the same tissue is the focus of both research efforts. These efforts may finally elucidate common molecular mechanisms that regulate adipogenesis and partitioning of lipids into different fat depots in humans, rodents and farm animal species.
Terms for description and methods of analysis of selected fat depots in different species.
| Term | Description | Human / Rodents | Farm Animals (Cattle, Pig) | References |
|---|---|---|---|---|
| Visceral adipose tissue (VAT) | Sum of cellular adipose depots in the peritoneal cavity (omental and perirenal fat covering digestive tract and kidney, respectively) | Computer tomography Sampling of omental fat during surgery possible | Quantitative dissection of omental and perirenal fat possible (post-mortem) | [3, 15, 16] |
| Subcutaneous adipose tissue (SAT) | Adipose tissue layer below the skin | Ultrasound measurement Sampling during surgery possible | Common term: subcutaneous fat (SCF), Ultrasound measurement, Sampling post-mortem or by biopsy technique | [15, 17, 18] |
| Deep subcutaneous fat (dSAT) | General term for inner layer of SAT at different locations | Macroscopic dissection, Biopsy technique | No investigations of different layers of SAT so far | [17] |
| Subfascical fat (SFAT) | Adipocytes located between inner layer of SAT and muscle | Computer tomography | Term not used | [16] |
| Intramyocellular lipids (IMCL) | Lipid droplets in the sarcoplasm of myocytes | Magnetic resonance spectroscopy, electron microscopy Histological staining (Oil Red-O; OsO4) | No separate measurement (see IMF) | [19] |
| Intramuscular fat (IMF) | Adipocytes located within a muscle | Term not used in human Described in rodents | Meat quality parameter Post-mortem chemical analysis (includes IMCL) Histological staining (Oil-Red-O) | [20, 21] |
| Marbling fat | Adipocytes located within a muscle | Term not used | Meat quality parameter Image analysis of visible groups of intramuscular adipocytes (post-mortem) | [15] |
| Extramyocellular lipids (EMCL) | Lipids (adipocytes?) located between myocytes | Magnetic resonance spectroscopy (human) Mostly not differentiated from IMCL | Term not used | [19] |
| Perimysial adipocytes | Adipocytes located within a muscle | Light and electron microscopy (rat) | Term not used | [22] |
| Intermuscular adipose tissue (IMAT) | Adipocytes located between muscles | Computer tomography | Post-mortem macroscopic dissection possible | [16, 20] |
Depot specific properties of adipose tissue
There are well established differences between VAT and SAT, in terms of cell morphology, metabolic activity and responsiveness, transcriptome and proteome (reviewed in [3, 27]). VAT is generally considered to be metabolically more active than SAT, with higher basal and insulin-stimulated glucose uptake and increased insulin metabolism [28]. Furthermore, it contains a larger proportion of inflammatory and immune cells and possesses a lesser preadipocyte differentiating capacity than SAT (reviewed in [27, 29]). Endocrine factors from VAT are secreted directly into the portal system with direct access to the liver, whereas proteins from SAT enter the systemic circulation. Consequently, adipose tissue should not be considered as a homogenous endocrine organ but rather as a group of organs with certain similarities and unique features according to their anatomical location [2]. Recent results confirmed this concept [17, 30, 31].
Humans and mice: Functional differences between VAT and SAT
Early investigations in humans analyzed single factors (e.g. adipsin, TNF-alpha, leptin) or transcriptomes from adipose tissue of mixed or non-specified origin, respectively [32-35]. Bolduc et al. (2004) [36] provided the first quantitative description of an adipose tissue transcriptome (retroperitoneal fat) in mice. Most of the following investigations aimed at detecting transcriptome differences between individuals in a single fat depot as a consequence of different physiological states or different treatments [37-48]. Zhang et al. (2007) [49] investigated inter-individual variability of gene expression in SAT and VAT of obese men. They described the adipose tissue transcriptome as relatively stable in general, however high inter-individual variation in gene sets linked to lipid metabolism and cell death was observed in both depots.
In contrast, fewer studies considered gene expression differences between adipose depots from different locations in the human body. Dolinkova et al. (2008) [50] analyzed 41 genes by RT-qPCR and found similar contributions of SAT and VAT to pro-inflammatory profiles in obese individuals. Whole transcriptome SAT and VAT gene expression levels of 75 obese individuals revealed an overlap of regulated inflammatory genes in both depots. The expression of these genes in SAT correlated with plasma HDL cholesterol, whereas their expression in VAT correlated with plasma glucose. This underlined the specific roles of VAT and SAT in glucose and lipid metabolism [51]. Del Pozo et al. (2011) [52] found striking differences in the number of regulated genes between VAT and SAT (1,268 vs. 74) of obese compared to lean subjects. This demonstrated that VAT reacted with more pronounced transcriptional changes to obesity compared with SAT. Fawcett et al. (2010) [28] described VAT as generally more active at metabolic level in comparison to other depots. Macotela et al. (2012) [53] observed large differences in adipogenic potential of precursor cells derived from either VAT or SAT and demonstrated that both cell populations require different signals for adipogenic differentiation. Adipogenic precursor cells (APCs) from VAT efficiently differentiated into mature adipocytes after induction by BMP4 and 2 (bone morphogenetic proteins 4 and 2) only, whereas cells from SAT did not depend on this signal. They conclude that VAT-derived APCs differ from SAT-derived cells in terms of gene expression signatures, growth factor requirements, and differentiation ability. VAT-derived APCs have more mesenchymal stem cell-like properties, expand by hypertrophy and contribute to inflammation in obesity. In contrast, subcutaneous adipose cells expand mainly by hyperplasia and stay metabolically intact in obesity. Furthermore, Tran et al. (2008) [54] conclude from results of heterologous fat transplantation in mice (SAT into VAT) that SAT possesses cell-autonomous properties that can act on other tissues to improve their metabolic state. Beside these intrinsic depot differences, recent results demonstrated the additional impact of epigenetic factors on adipose tissue expression profiles. Autologous transplantation of VAT with low expression of adiponectin, leptin, resistin, and visfatin to subcutaneous sites of rats with diet-induced obesity resulted in an increase of the expression of these genes to levels typical for SAT. This suggested that local factors (e.g. glucose) of their new anatomical location modified the expression of adipokines by recruitment of key enzymes for histone methylation [55].
Humans and mice: Body location and sex determine properties of adipose tissue
Yamamoto et al. (2010) [30] investigated the expression of developmental genes and adipogenic transcription factors in different murine depots of white adipose and brown adipose tissues, respectively. They detected depot-specific developmental signatures with Shox2 (short stature homeobox 2) expression specific for SAT compared to different VAT depots. Furthermore, increased Tbx15 (T-box 15) and En1 (engrailed 1) expression was related to the dorsal-ventral distribution of SAT. These results indicated specific signatures even for SAT located in different regions of the body. All of the detected differences were largely independent of nutritional state and revealed a depot-specific developmental program of adipocyte differentiation and function.
Macotela et al. (2009) [56] reported an important role of sex and sex steroids on functional properties of adipocytes from different depots. VAT adipocytes from female mice had increased lipogenic capacity as well as increased lipolytic rates. The resulting higher metabolic turnover of adipocytes in females would explain their lower size compared to those from males. Furthermore, female VAT-derived adipocytes were more insulin-sensitive than those derived from female SAT and from both depots in males. Circulating estrogen at high levels reduced both lipogenic rates and depot size preferentially in VAT. In contrast, testosterone inhibited lipogenesis and decreased insulin sensitivity in both depots of male mice [56]. Recent genome wide association analyses revealed a sex specific locus for total body fat content in men (IRS1; insulin receptor substrate 1; [57]) which was further confirmed by Fox et al. (2012) [58]. The latter authors identified a SNP at the human THNSL2 (threonine synthase-like 2) locus that was significantly associated with the amount of VAT in women but not in men. These results provide further evidence that VAT and SAT differ in numerous physiological parameters which are caused by specific genetic architecture and epigenetic modifications.
Farm animals: Functional properties of VAT, SAT, IMAT, and IMF
In contrast to humans and rodents, investigations of visceral fat depots in farm animals are rather scarce. These studies either targeted expression of single adipokines and transcription factors [59, 60] or aimed at comparing expression profiles of non-adipose tissues [61]. In a differential display approach, six genes were identified with expression differences in bovine VAT and SAT. All of these differences were reproducible in swine and mice, indicating that depot-specific factors exist over species [62]. Li et al. (2012a, b) [63, 64] comparatively analyzed whole genome gene expression and methylation in SAT from three different locations, four VAT depots and intermuscular fat in swine, respectively. They detected depot specific, co-methylated gene sets which linked different SAT depots to metabolic pathways, whereas co-methylated genes in VAT depots affected serum levels of inflammatory and immune adipokines like IL6 (interleukin 6), TNFA (tumor necrosis factor alpha) and SERPINE1 (serpin peptidase inhibitor, clade E member 1; [63]). Most interestingly, intermuscular adipose tissue (IMAT) was characterized by a similar co-methylation signature like that found in VAT supporting the notion that IMAT may constitute an independent risk factor for metabolic diseases in human [65].
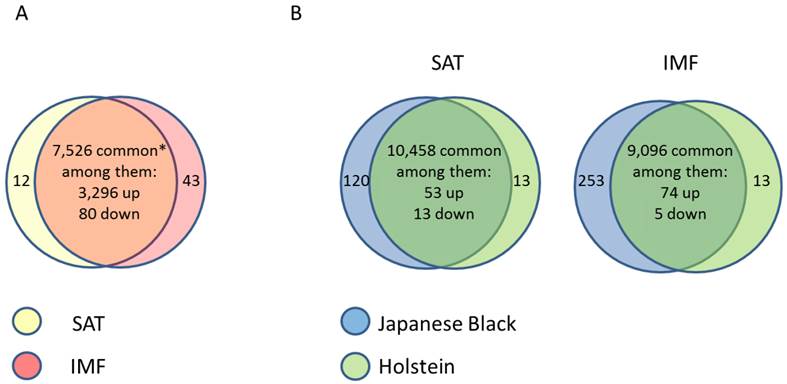
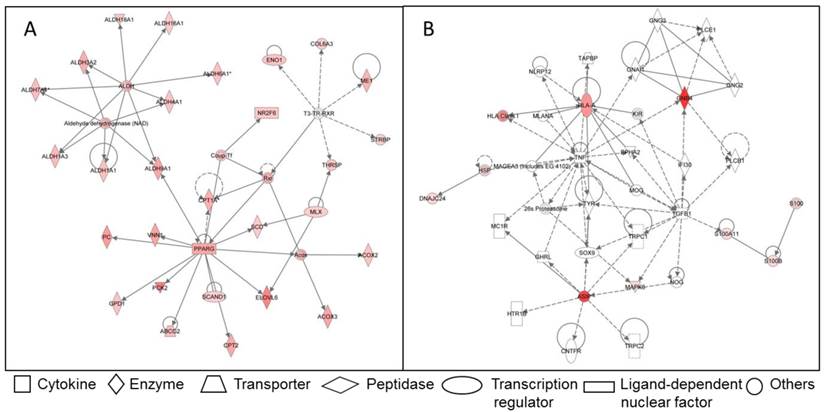
Due to the importance of IMF for meat quality, several studies targeted the IMF transcriptome in farm animal species. Bong et al. (2010) [66] compared the expression profiles of bovine SAT and IMF in Korean native cattle using serial analysis of gene expression (SAGE). They identified 50 and 32 genes that were more than 2-fold up- and down-regulated in SAT compared to IMF, respectively. In an own experiment [67], global analyses with GeneChip® Bovine Genome Array (Affymetrix, USA) in Japanese Black and Holstein steers revealed almost 3,400 genes that were differentially expressed between SAT and IMF independent of breed (Figure 1A). This is much greater than the number of differentially expressed genes within an adipose depot between both divergent breeds (IMF: 66; SCF: 79; Figure 1B; [68]). The vast majority of identified genes were down-regulated in IMF, probably indicating a lower transcriptional activity of adipocytes embedded in skeletal muscle compared to SAT. Several genes up-regulated in SAT [66] were confirmed by Albrecht et al. (2010) [68] (e.g. ANXA5, annexin A5; SPARC, osteonectin; FTH1, ferritin, heavy polypeptide 1, and DGAT2, diacylglycerol O-acyltransferase 2). The large differences in gene expression between SAT and IMF are reflected in the respective networks of regulated genes (Figure 2). The depot comparison reveals large, closely interconnected groups of genes with potential direct interactions (Figure 2 A). This is in clear contrast to the differences between breeds within a depot (IMF), with few related genes and rather spurious relationships (Figure 2 B; [68]). The networks indicate that lipid metabolism is more divergent between adipose depots within an animal than between meat- and milk-type cattle breeds. Similar results were reported by Zhou et al. (2010) [69] who compared cultured porcine subcutaneous and intramuscular adipocytes. These data underline the heterogeneity of VAT, SAT and muscle-related adipose depots at the transcriptional level independent of species. Different cell fractions within adipose tissue may contribute in a specific manner to the transcriptome of the tissue as whole. Variation in the composition of the depots is a likely reason for that [50] beside depot-specific expression profiles of adipocytes, preadipocytes and other cells [70, 71]. This is supported by results of Ortiz-Colon et al. (2009) [72] who observed a different adipogenic capacity of stromal vascular cells derived from bovine VAT, SAT and IMF in decreasing order.
(A) Comparison of gene expression profiles between subcutaneous (SAT, n = 6) and intramuscular (IMF, n = 6) adipose tissue of Japanese Black (n = 3) and Holstein (n = 3) cattle (between depots over breeds). * Only transcripts without breed differences were included. Approximately 45 % of transcripts were differently regulated between the depots regardless of breed. (B) Comparison of gene expression profiles in SAT and IMF derived from Japanese Black (n = 3) and Holstein (n = 3) cattle (between breeds within depot). Only 0.6 % (SAT) and 0.9 % (IMF) were differently regulated between the breeds within the respective depots [68]. Genes were considered as differently expressed with fold change > 2.0, p < 0.05 and q < 0.30.
Examples for networks of differently regulated genes in the functional category “lipid metabolism” in adipose tissue of Japanese Black and Holstein cattle. (A) Network of genes up-regulated in SAT compared to IMF regardless of breed. Only genes without breed differences were included. (B) Network of genes up-regulated in IMF of Japanese Black compared to IMF of Holstein cattle. Up-regulated genes are marked reddish/red. Solid lines indicate direct interaction between genes, dashed lines stand for indirect interactions [68].
VAT depots in meat-producing farm animals (e.g. omental, perirenal and intestinal fat) are mostly discarded and considered as “waste fat”. Since there exist large breed differences in accretion of VAT depots under similar feeding conditions (e.g. cattle; [73]), metabolic consequences of these differences should be investigated. VAT must be understood as part of the animals' endocrine system in the future. Parameters describing animal fat depots quantitatively and qualitatively may be integrated in the description of phenotypes, and finally constitute new selection criteria.
Proteomes and secretomes of adipose tissue depots
Due to post-transcriptional processes, the relationship between transcript abundance and the amount of biologically active proteins is not always very close and consequently transcriptome analyses should be complemented by proteome studies [74]. Own investigations on a single adipokine (agouti encoded by the ASIP gene) in different bovine adipose tissues did not reveal an effect of transposon-mediated mRNA over-expression on protein abundance and cellular localization thus underlining the necessity of this combined approach [75]. Recent advances in global proteomic technologies has led to a growing number of respective analyses of cultured cells and adipose tissue published in the last decade (reviewed in [3]). The review lists 65 studies on human and animal adipose proteomes and an additional 3 investigations that have been published regarding human and swine since then [76-78]. However, only 6 of 68 analyses targeted differences between fat depots (VAT vs. SAT or IMF), and in part between their components (stromal vascular fraction vs. mature adipocytes), in human, rat, cattle and pig. All of these studies confirmed different protein profiles depending on anatomical location and implied different functionality of the adipose depots [79-84]. Most of the proteins which were enriched in the VAT proteomes compared to SAT were related to stress response and inflammation in human as well as in cattle [80, 83]. The proteomic data largely confirmed the notion of VAT being metabolically more active than SAT. Proteins expressed in adipose tissue components may either constitute structural and functional elements of the cells or they are secreted via different ways. The latter group is of major interest as their members can act on other cells in an auto-, para- or endocrine manner [85].
Trayhurn & Wood (2004) [86] suggested the term “adipokines” for proteins secreted by adipose tissue. For sake of clarity, this term was later restricted to proteins secreted by adipocytes rather than by other cells in the tissue (i.e. macrophages) [85]. Considering the large number of adipokines detected or predicted so far (> 600 [5]), their totality is called “adipokinome” or “secretome”. Alvarez-Llamas et al. (2007) [87] characterized the secretome of human VAT explants and identified 108 secreted proteins. The functional categories with the highest number of proteins were “extracellular matrix”, “signaling/regulatory function”, “involved in degradation”, and “immune function”. Zvonic et al. (2007) [88] detected more than 100 proteins secreted from human adipose-derived stem cells. Their major finding was the modulation of expression of the serpin family of protease inhibitors during the course of adipogenesis. Dynamic changes of the adipocyte secretome during adipogenesis were also confirmed by other studies [89-91]. A large-scale characterization of conditioned medium derived from differentiated human subcutaneous adipocytes resulted in 263 potentially secreted proteins, including 44 proteins that were not previously described. The overlap of identified proteins with those from earlier studies was between 36 and 57% and indicated that the human adipokinomes are still incompletely characterized [76].
A comparison of the SAT secretome described by Lehr et al. (2012) [76] with mRNA expression profiles of VAT and SAT from lean and obese subjects demonstrated that ~65% of the respective transcripts were regulated by obesity and ~55% were differently expressed in VAT and SAT, respectively [92]. Four genes were regulated by obesity and depot and hence considered as promising candidates for further investigations (WNT1-inducible-signaling pathway protein 2, WISP2; transmembrane glycoprotein NMB, GPNMB; inter-alpha-trypsin inhibitor heavy chain H5, ITIH5; and complement C4-A, C4A). Recently, Taube et al. (2012) [93] demonstrated the role of adipokines in mediating lipotoxic effects of palmitic acid on skeletal muscle cells. Low doses of palmitic acid exerted lipotoxicity when incubated with conditioned medium derived from cultured subcutaneous adipocytes. This effect could not be counteracted by muscle contraction. The authors concluded that conditioned medium mimics the complex secretome of adipocytes and many factors contribute to the effect on skeletal muscle. This study constitutes a conclusive example for cross-talk between peripheral adipose depots and skeletal muscle.
Investigations in farm animals targeting the secretome of adipose depots have not been undertaken yet. One major task in the future will be unraveling the function of known adipokines [85], detecting new adipokines, and understanding the physiological role of adipokinomes from adipose tissue depots in different species.
Taking into consideration that also skeletal muscle tissue is secreting proteins with endocrine function, the spectrum of potential direct communication between skeletal muscle and adipose tissue is considerably widened. The following sections thus will summarize the current knowledge on these secreted proteins, the so-called myokines, and discuss potential implications for adipose-muscle cross-talk.
Myokines - an emerging new class of secreted proteins
Although myostatin was identified as a protein secreted from muscle cells with crucial importance for skeletal muscle development in 1997 [94, 95], the perception of skeletal muscle as endocrine organ gained acceptance several years later [96]. Early research on proteins secreted from myocytes focused mainly on interleukins (IL), namely IL-6, IL-8 and IL-15 (reviewed in [97]). The classical and best described member of this family is IL-6. When it was discovered that levels of IL-6 in plasma were positively related to duration of physical exercise in human, the secretory role of contracting muscle was unveiled [98].
Humans: Skeletal muscle cells secrete a large number of proteins upon contraction
Pedersen et al. (2003) [99] coined the term “myokines” for IL-6 and other cytokines, which are produced and released by skeletal muscle and exert their effects in other organs. Trayhurn et al. [85] suggested the use of this term for all muscle-secreted proteins regardless of the fact whether they are related to the immune system (cytokines) or not. Until now, a limited number of myokines have been characterized. Recently, an additional member of the IL-family (IL-7) has been described. Moreover, brain-derived neurotrophic factor (BDNF; reviewed in [100]), leukemia inhibitory factor (LIF; [101]), oncostatin M (OSM; [102]) chemokine CXC motif ligand 1 (CXCL-1; [103]) and myonectin (CTRP15; [104]) have also been described. Beside effects on inflammatory processes and immune function, myokines are involved in myocyte proliferation, inhibition of oncogenic processes and systemic lipid metabolism [85, 102, 104]. An intrinsic property of many myokines is the dependence of their secretion on skeletal muscle contraction. By this, a link between physical activity and beneficial health effects could be established. Recently, human muscle cell culture models have been developed, in which electrical pulse stimulation triggers contraction [105, 106]. This provides an excellent in vitro-model to elucidate contraction-induced myokines. Recent advances in proteomics have allowed for more comprehensive analyses of proteins secreted from skeletal muscle. Several studies targeting muscle cells from human, rat and mouse led to identification of several hundreds of secreted proteins belonging to different functional categories with many being regulated during myogenesis (reviewed in [85]). Considering this large number, the term “myokinome” was introduced for the totality of secreted proteins from myocytes [85].
Humans and mice: Myokines are involved in cross-talk with other tissues
Henningsen et al. (2010) [107] identified 635 putative secreted proteins from murine C2C12 cells and quantitatively profiled 624 of them at days 0, 2, and 5 of differentiation. As previously noted for differentiating adipocytes [89-91], the myokinome was also highly dynamic with 30% of secreted proteins differentially regulated during myogenesis. “Extracellular matrix structural component” and “cytokine activity” were among the highest enriched functional clusters, thus linking the myokinome and the adipokinome [87, 107]. The rather low relationship between mRNA and protein dynamics of selected members of the IGF- (insulin-like growth factor), TGF- (transforming growths factor) and CC chemokine- (chemokine [C-C motif] ligand) families in that study indicated post-transcriptional and post-translational mechanisms as causes for the differential release from murine muscle [107, 108]. The observed expression of CC chemokines (MCP-1, 2, 3; monocyte chemotactic proteins 1, 2 and 3) upon pro-inflammatory stimuli was considered as sign of involvement of muscle in a broader cross-talk with inflammatory-dependent systems [108]. MCP-1 exerts specific insulin-resistance inducing effects in adipocytes as well as in skeletal muscle in vitro and is expressed in both tissues. This makes it likely that MCP-1 actively participates in the cross-communication between both tissues (reviewed in [85]).
Only recently, a transcriptional co-activator was identified in mice (PGC-1α encoded by PPARGC1A, peroxisome proliferator-activated receptor gamma, coactivator 1 alpha) that activates a hitherto unknown myokine with major impact on adipose tissue physiology [109]. In this study cleavage of the membrane protein FNDC5 (fibronectin type III domain containing 5) and secretion of a part of it upon stimulation by increased PGC-1α expression was demonstrated. The secreted protein, called irisin, constitutes a new myokine which induces “browning” of white adipose tissue and subsequent thermogenesis. Irisin is assumed to mediate its function by an unidentified cell surface receptor. Mormeneo et al. (2012) [110] described a mitochondrial and myokine transcriptional program induced by PGC-1α over-expression in human skeletal muscle cell culture that probably links this co-activator to physical exercise and inflammation. Furthermore, different mechanisms involving TGFß- (transforming growth factor beta) signaling and ALDH1A1 (aldehyde dehydrogenase 1 family, member A1) were detected which increased the expression of PGC-1α in murine adipose tissue and induced a thermogenic program [111, 112]. Mice lacking PGC-1α in adipose tissue developed insulin resistance when fed a high-fat diet [113]. PGC-1α is considered as a master regulator of mitochondrial biogenesis and oxidative metabolism in several cell types including brown fat and skeletal muscle but not white adipose tissue (reviewed in [114]). Lin et al. (2002) [115] described a preferential expression of PGC-1α in slow-twitch muscle fibers which rely more on oxidative metabolism. This makes differential expression of PGC-1α in skeletal muscles with different fiber type compositions likely and may modulate the cross-talk with adipose tissue. Furthermore, PGC-1α was identified as essential co-activator of PRDM16 (PR domain containing 16) and is thus involved in the brown fat genetic program via an additional pathway [114]. There is growing evidence that PRDM16 is able to induce a brown fat transcriptome in brown preadipocytes, myogenic cells, fibroblasts, and upon transgenic expression in white adipose tissue [116-119]. The involvement of BMP7 (bone morphogenetic protein 7), CEBPB, and microRNAs in cell fate determination and trans-differentiation of adipocytes underlines the complexity of this process but opens numerous potential intervention strategies to fight obesity in men [118, 120, 121]. PGC-1α as a central molecule in these pathways exerts additional influence by induction of myokine secretion and thus, links adipose tissue with skeletal muscle.
Farm animals: Skeletal muscle proteomes
The first proteomic analyses of skeletal muscle in farm animals aimed mainly at meat quality and consequently considered muscle tissue after slaughter [122]. Meanwhile, numerous studies have been published comparing skeletal muscle proteomes in pig and cattle. They were predominantly directed on identification of key factors for meat quality variations (e.g. [123-125]), but also compared effects of different muscle fiber type composition [126], compensatory growth [127], genetically determined muscle hypertrophy [128], intrauterine growth retardation [129], and ontogenetic stage [130]. All of these studies considered the total proteome without differentiating between structural and functional components of the muscle cell itself on one hand and secreted proteins, on the other hand. For this reason, data on farm animal myokinomes do not exist to date. The results recently reported in humans and rodents however, may initiate targeted approaches for analyzing factors like i.e. PRDM16 and PGC-1α in meat producing animals at the protein and functional levels.
The PPARGC1A locus, encoding PGC-1α, was studied in numerous association analyses with fatness traits in farm animals. The results however, are contradictory. Significant relationships of PPARGC1A polymorphisms with milk fat composition and backfat thickness in cattle have been reported [131, 132] which were partly in conflict with earlier reports [133, 134]. In contrast, investigations in swine revealed significant effects of nucleotide variation at the PPARGC1A locus on muscle fiber characteristics and meat quality traits (e.g. [135-137]. Erkens et al. (2008) [138] described two novel splice variants of porcine PPARGC1A that were also detected in human tissue. Only recently a relationship between body weight in cattle and polymorphisms of the PRDM16 locus was established [139]. Although the studies did not investigate these factors at protein level, the results collectively indicated that farm animals may serve as models to investigate the effect of genetic variants on protein function and subsequently on phenotypes. Since muscle contraction is of crucial importance for the secretion of myokines, these findings may in turn add a new dimension to the evaluation of animal husbandry systems (e.g. pasture vs. confined housing).
Muscle associated fat - a special role in cross-talk with muscle?
Humans and mice: IMCL and IMAT - underestimated adipose depots so far?
As mentioned before, VAT and SAT have been the focus of research in human and rodents with the general aim to link adipose depot physiology to pathological conditions in human. However, increased intramyocellular lipid (IMCL) deposition was also related to type 2 diabetes mellitus (T2DM) and insulin resistance in human [19, 140]. A genetic predisposition was assumed to cause a reduced ability to oxidize skeletal muscle fatty acids. This might explain the sustained insulin resistance in affected subjects even after weight loss and physical exercise [141]. On the other hand, endurance trained athletes showed a marked increase in IMCL while skeletal muscle remained insulin sensitive [16, 142, 143]. Intramyocellular lipid droplets are now recognized as biologically active organelles rather than as inert stores of lipids (reviewed in [144]). A proteomic approach revealed 324 lipid droplet-associated proteins with many of them belonging to functional categories “lipid metabolism”, “membrane traffic” and “mitochondrial proteins” [145].
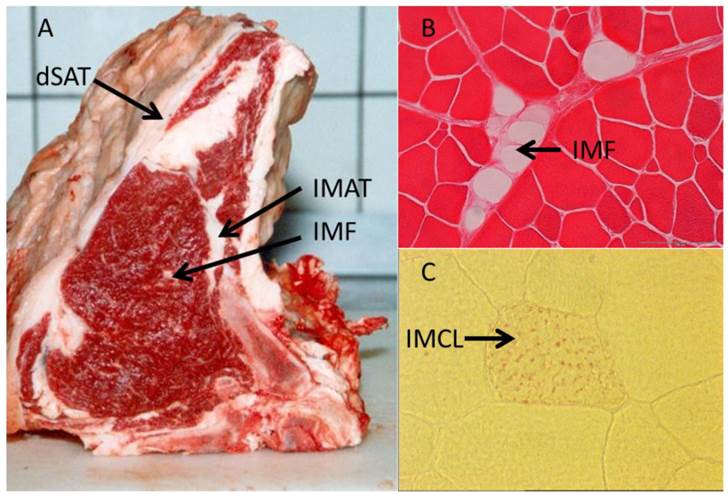
In contrast to IMCL located within the myocytes, IMAT and IMF appear as cellular structures between muscles and within muscles, respectively (Figure 3 and Table 1). Goodpaster et al. (2000) [16] revealed a positive correlation between the amount of adipose tissue interspersed around and between skeletal muscle and insulin resistance. In contrast, no similar relationship was found between SAT and insulin sensitivity in the same individuals. This indicated peculiarities of muscle-associated adipose tissue compared to other depots. Yim et al. (2007) [65] investigated individuals of different ethnic groups and sex and found significant correlations between IMAT amount and cardiovascular risk factors independent from that mediated by other fat depots. In that study, IMAT reached 28 - 82 % of the mass of VAT. Consequently, this and further studies considered IMAT as a depot of similar size and importance for metabolism like VAT [65, 146]. While the latter authors discussed a more passive role of IMAT in influencing e.g. the glucose metabolism of the muscle by impairing blood flow, Vettor et al. (2009) [147] suggested a reciprocal communication mediated by biologically active molecules - adipokines and myokines. They reported recruitment of muscle stem cells into the adipogenic lineage as a reaction to a primary insult. High glucose levels were identified as one possible trigger for this process in cultured rat stem cells [148]. Molecular mechanisms involved in this conversion include, among others, peroxisome proliferator-activated receptors (PPARs) as well as muscle derived factors like myostatin, strengthening the hypothesis of a bi-directional communication between (pre) adipocytes and muscle cells (reviewed in [147]).
To date, only the secretomes of human adipocytes have been profiled from commercial cell lines or primary SAT/VAT derived cell cultures [76]. Results for muscle associated adipose tissue are not yet available. Further investigations are necessary to elucidate the potential role of muscle-associated fat depots in muscle-fat cross-talk via paracrine mechanisms.
Illustration of different muscle-related adipose tissue depots. (A) Deep subcutaneous adipose tissue (dSAT) covering M. serratus dorsalis, intermuscular adipose tissue (IMAT) between M. intercostalis interni and M. longissimus dorsi, and intramuscular fat (IMF) within M. longissimus dorsi in cattle. (B) Cellular structure of IMF in M. longissimus dorsi (cattle, Eosin stained). (C) Intramyocellular lipids (IMCL, red dots, Oil-red O stained) in a muscle cell (M. longissimus dorsi, mouse) [unpublished].
Humans: Adipo-myokines as mediators of cross talk
In a recent review, Trayhurn et al. (2011) [85] noted that several proteins identified as adipokines are also components of the myokinome. Some of these proteins seem to have different functions depending on the physiological situation. IL-6 as a myokine is of critical importance for muscle contraction, whereas chronic elevation of the adipokine IL-6 may induce muscle insulin resistance. Therefore, the term “adipo-myokine” might be appropriate for IL-6 and further factors like members of the interleukin family and MCP-1 [85].
Scheele et al. (2012) [149] reported striking differences in IL-6 responsiveness of satellite cells derived from skeletal muscle of healthy, obese, or obese subjects with T2DM, respectively. They found that IL-6 signaling was negatively controlled in muscle of obese persons by down-regulation of the IL-6Rα (interleukin receptor alpha), and that dysfunctional IL-6 signaling was established further downstream of the IL-6 receptor in T2DM subjects. The permanently increased levels of IL-6 in obesity and T2DM may result from a compensatory attempt to induce more insulin. As a side effect, IL-6 signaling in skeletal muscle is then impaired [149]. Whether muscle associated fat is the major source of IL-6 remains to be elucidated.
Mice: Heterogenous characteristics of intramuscular adipocyte progenitors
Genetic variation in IMF content of mice was similar to that seen in swine and thus, quantitative trait loci for this parameter in M. longissimus were mapped [150]. Functional investigations revealed intrinsic specificities of intramuscular adipocytes compared to those from other depots as well as indications for further subdivision of IMF adipocytes and progenitor cells. Liu et al. (2012) [21] found a higher adipogenic capacity in predominantly slow, oxidative soleus muscle than in fast glycolytic M. extensor digitorum of mice. This was obviously caused by a higher proportion of adipocyte progenitor cells in slow M. soleus but also by intrinsically different growth and differentiation properties of the adipose progenitor cells in different muscles. Joe et al. (2009) [151] demonstrated similar differences between adipogenic progenitors of VAT and SAT in mice leading to different responsiveness of the depots to a high-fat diet in mice. Additionally, intramuscular adipogenic progenitors are able to facilitate myogenesis by expansion thus supplying signals - most likely promyogenic cytokines - to myogenic progenitors. Notably, these adipogenic progenitors do not generate myofibers themselves [152]. Interestingly, these muscle-resident adipogenic progenitors are not derived from satellite cells or Pax3-(paired box 3) expressing mesodermal cells but likely descend from haematopoietic or mesenchymal lineages [21, 153, 154]. The different descendence and the involvement of intramuscular adipocytes in skeletal muscle regeneration define intramuscular adipocytes as a special population which itself may be heterogeneous. Furthermore, these results provide evidence for paracrine effects of IMF on surrounding skeletal muscle. Reciprocal signaling from myocytes to adipocytes is very likely. This implies that co-culture of skeletal muscle cells with skeletal muscle-derived adipocytes is a suitable technique for further elucidation of crosstalk. Laser microdissection might complement this approach by allowing separation of adipocytes from myocytes in their “natural” environment with subsequent transcript analyses [155].
Farm animals: IMAT and IMF - depots with impact on meat quality
Intermuscular fat (IMAT) in farm animals is perceived as a non-dissectible component of meat for human consumption and thus, was characterized in this respect only. IMAT had the lowest lipid content and the smallest adipocyte size compared to porcine SAT and VAT, and was considered as a depot with unique properties (reviewed in [156]).
In contrast, IMF - as a determinant of the eating quality of meat - attracted more attention in farm animals. In contrast to numerous studies on gene expression (see above), only few targeted the proteome.
Gondret et al. (2008) [79] identified and characterized 15 proteins with significantly different abundance in porcine IMF compared to other adipose depots. As most of these proteins were down-regulated, a lower lipogenic ability and reduced β-oxidation of intramuscular adipocytes was assumed, compared to cells derived from other depots. Since they initially detected almost 150 proteins considered as characteristic for IMF, the real number of proteins specific for intramuscular adipocytes should be much higher. However, the identification of proteins potentially secreted from IMF in farm animals is still pending. Although it is not clear whether IMF in farm animals constitutes an adipocyte population different from IMAT, it is very likely that IMF exerts similar metabolic effects on surrounding muscle cells like IMAT and a reciprocal influence of myocytes on intramuscular adipocytes might exist. Understanding basic principles of this bi-directional communication would greatly enhance the opportunities to influence carcass composition in farm animals.
Summary
Adipose tissue and skeletal muscle are increasingly perceived as major endocrine organs. The application of advanced genomic and proteomic technologies allowed the identification and - to a much smaller extent - the characterization of expressed genes and proteins. A large number of proteins are secreted and exert endo-, para- and autocrine effects, thus modulating the body composition in humans, mice and farm animals. The totality of the secreted proteins is called adipokinome and myokinome, respectively. There is growing evidence for depot-specific properties of adipocytes as well as for specificities of differently composed muscles. Due to their close proximity to muscle cells, intra- and intermuscular adipocytes may exert special influence on skeletal muscle and vice versa, are subject to regulation by myokines. The complexity of the adipose-muscle crosstalk becomes obvious when one keeps in mind that this communication is embedded in larger regulatory networks involving organs like liver and nervous system. Better understanding of the complex cross-talk between intramuscular adipocytes and skeletal muscle will give rise to strategies to improve human health as well as for sustainable production of high quality meat.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337-350
2. Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab. 2004;89:2548-2556
3. Peinado JR, Pardo M, de la Rosa O. et al. Proteomic characterization of adipose tissue constituents, a necessary step for understanding adipose tissue complexity. Proteomics. 2012;12:607-620
4. Ahima RS. Adipose tissue as an endocrine organ. Obesity (Silver Spring). 2006;14(Suppl 5):242S-249S
5. Lehr S, Hartwig S, Sell H. Adipokines: A treasure trove for the discovery of biomarkers for metabolic disorders. Proteomics Clin Appl. 2012;6:91-101
6. Harwood HJ Jr. The adipocyte as an endocrine organ in the regulation of metabolic homeostasis. Neuropharmacology. 2012;63:57-75
7. Conde J, Scotece M, Gomez R. et al. Adipokines: biofactors from white adipose tissue. A complex hub among inflammation, metabolism, and immunity. Biofactors. 2011;37:413-420
8. Tersigni C, Di NF, D'Ippolito S. et al. Adipokines: new emerging roles in fertility and reproduction. Obstet Gynecol Surv. 2011;66:47-63
9. Deng Y, Scherer PE. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann N Y Acad Sci. 2010;1212:E1-E19
10. Weiss TW, Seljeflot I, Hjerkinn EM. et al. Adipose tissue pro-inflammatory gene expression is associated with cardiovascular disease. Int J Clin Pract. 2011;65:939-944
11. Gerdes S, Rostami-Yazdi M, Mrowietz U. Adipokines and psoriasis. Exp Dermatol. 2011;20:81-87
12. Gustafson DR. Adiposity hormones and dementia. J Neurol Sci. 2010;299:30-34
13. Wajchenberg BL, Giannella-Neto D, da Silva ME. et al. Depot-specific hormonal characteristics of subcutaneous and visceral adipose tissue and their relation to the metabolic syndrome. Horm Metab Res. 2002;34:616-621
14. Allen CE. Cellularity of adipose tissue in meat animals. Fed Proc. 1976;35:2302-2307
15. Albrecht E, Teuscher F, Ender K. et al. Growth- and breed-related changes of marbling characteristics in cattle. J Anim Sci. 2006;84:1067-1075
16. Goodpaster BH, Thaete FL, Kelley DE. Thigh adipose tissue distribution is associated with insulin resistance in obesity and in type 2 diabetes mellitus. Am J Clin Nutr. 2000;71:885-892
17. Walker GE, Verti B, Marzullo P. et al. Deep subcutaneous adipose tissue: a distinct abdominal adipose depot. Obesity (Silver Spring). 2007;15:1933-1943
18. Leahy S, Toomey C, McCreesh K. et al. Ultrasound measurement of subcutaneous adipose tissue thickness accurately predicts total and segmental body fat of young adults. Ultrasound Med Biol. 2012;38:28-34
19. Jacob S, Machann J, Rett K. et al. Association of increased intramyocellular lipid content with insulin resistance in lean nondiabetic offspring of type 2 diabetic subjects. Diabetes. 1999;48:1113-1119
20. Hoshino T, Suzuki A, Yamaguchi T. et al. A Comparative Morphometrical Analysis of the Amount and Distribution of Fat within Muscles of Japanese Black Cattle, Japanese Shorthorn, and their Crossbreed (F1) Steers. Tohoku Journal of Agriculture Research. 1990;40:57-64
21. Liu W, Liu Y, Lai X. et al. Intramuscular adipose is derived from a non-Pax3 lineage and required for efficient regeneration of skeletal muscles. Dev Biol. 2012;361:27-38
22. Belmonte MA, Aoki MS, Tavares FL. et al. Rat myocellular and perimysial intramuscular triacylglycerol: a histological approach. Med Sci Sports Exerc. 2004;36:60-67
23. Morlein D, Link G, Werner C. et al. Suitability of three commercially produced pig breeds in Germany for a meat quality program with emphasis on drip loss and eating quality. Meat Sci. 2007;77:504-511
24. Hausman GJ, Dodson MV, Ajuwon K. et al. Board-invited review: the biology and regulation of preadipocytes and adipocytes in meat animals. J Anim Sci. 2009;87:1218-1246
25. Paulke T, Pfuhl R, Maak S. Rohnährstoffgehalt und sensorische Eigenschaften des M. longissimus verschiedener deutscher Schweineherkünfte. Züchtungskunde. 2011;83:415-425
26. Dodson MV, Jiang Z, Chen J. et al. Allied industry approaches to alter intramuscular fat content and composition in beef animals. J Food Sci. 2010;75:R1-R8
27. Ibrahim MM. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes Rev. 2010;11:11-18
28. Fawcett J, Sang H, Permana PA. et al. Insulin metabolism in human adipocytes from subcutaneous and visceral depots. Biochem Biophys Res Commun. 2010;402:762-766
29. Tran TT, Kahn CR. Transplantation of adipose tissue and stem cells: role in metabolism and disease. Nat Rev Endocrinol. 2010;6:195-213
30. Yamamoto Y, Gesta S, Lee KY. et al. Adipose depots possess unique developmental gene signatures. Obesity (Silver Spring). 2010;18:872-878
31. Alvehus M, Buren J, Sjostrom M. et al. The human visceral fat depot has a unique inflammatory profile. Obesity (Silver Spring). 2010;18:879-883
32. White RT, Damm D, Hancock N. et al. Human adipsin is identical to complement factor D and is expressed at high levels in adipose tissue. J Biol Chem. 1992;267:9210-9213
33. Hotamisligil GS, Arner P, Caro JF. et al. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest. 1995;95:2409-2415
34. Zhang Y, Proenca R, Maffei M. et al. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425-432
35. Maeda K, Okubo K, Shimomura I. et al. Analysis of an expression profile of genes in the human adipose tissue. Gene. 1997;190:227-235
36. Bolduc C, Larose M, Lafond N. et al. Adipose tissue transcriptome by serial analysis of gene expression. Obes Res. 2004;12:750-757
37. Gomez-Ambrosi J, Catalan V, Diez-Caballero A. et al. Gene expression profile of omental adipose tissue in human obesity. FASEB J. 2004;18:215-217
38. Higami Y, Pugh TD, Page GP. et al. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415-417
39. Li J, Yu X, Pan W. et al. Gene expression profile of rat adipose tissue at the onset of high-fat-diet obesity. Am J Physiol Endocrinol Metab. 2002;282:E1334-E1341
40. Zhang J, He Q, Liu QY. et al. Differential gene expression profile in pig adipose tissue treated with/without clenbuterol. BMC Genomics. 2007;8:433
41. Pilvi TK, Storvik M, Louhelainen M. et al. Effect of dietary calcium and dairy proteins on the adipose tissue gene expression profile in diet-induced obesity. J Nutrigenet Nutrigenomics. 2008;1:240-251
42. van Dijk SJ, Feskens EJ, Bos MB. et al. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am J Clin Nutr. 2009;90:1656-1664
43. Shea J, French CR, Bishop J. et al. Changes in the transcriptome of abdominal subcutaneous adipose tissue in response to short-term overfeeding in lean and obese men. Am J Clin Nutr. 2009;89:407-415
44. Bol VV, Delattre AI, Reusens B. et al. Forced catch-up growth after fetal protein restriction alters the adipose tissue gene expression program leading to obesity in adult mice. Am J Physiol Regul Integr Comp Physiol. 2009;297:R291-R299
45. Marquez-Quinones A, Mutch DM, Debard C. et al. Adipose tissue transcriptome reflects variations between subjects with continued weight loss and subjects regaining weight 6 mo after caloric restriction independent of energy intake. Am J Clin Nutr. 2010;92:975-984
46. Grant RW, Vester Boler BM, Ridge TK. et al. Adipose tissue transcriptome changes during obesity development in female dogs. Physiol Genomics. 2011;43:295-307
47. Morton NM, Nelson YB, Michailidou Z. et al. A stratified transcriptomics analysis of polygenic fat and lean mouse adipose tissues identifies novel candidate obesity genes. PLoS One. 2011;6:e23944
48. Jin W, Olson EN, Moore SS. et al. Transcriptome analysis of subcutaneous adipose tissues in beef cattle using 3' digital gene expression-tag profiling. J Anim Sci. 2012;90:171-183
49. Zhang Y, Bosse Y, Marceau P. et al. Gene expression variability in subcutaneous and omental adipose tissue of obese men. Gene Expr. 2007;14:35-46
50. Dolinkova M, Dostalova I, Lacinova Z. et al. The endocrine profile of subcutaneous and visceral adipose tissue of obese patients. Mol Cell Endocrinol. 2008;291:63-70
51. Wolfs MG, Rensen SS, Bruin-Van Dijk EJ. et al. Co-expressed immune and metabolic genes in visceral and subcutaneous adipose tissue from severely obese individuals are associated with plasma HDL and glucose levels: a microarray study. BMC Med Genomics. 2010;3:34
52. del Pozo CH, Calvo RM, Vesperinas-Garcia G. et al. Expression profile in omental and subcutaneous adipose tissue from lean and obese subjects. Repression of lipolytic and lipogenic genes. Obes Surg. 2011;21:633-643
53. Macotela Y, Emanuelli B, Mori MA. et al. Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes. 2012;61:1691-1699
54. Tran TT, Yamamoto Y, Gesta S. et al. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 2008;7:410-420
55. Satoor SN, Puranik AS, Kumar S. et al. Location, location, location: Beneficial effects of autologous fat transplantation. Sci Rep. 2011;1:81
56. Macotela Y, Boucher J, Tran TT. et al. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes. 2009;58:803-812
57. Kilpelainen TO, Zillikens MC, Stancakova A. et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753-760
58. Fox CS, Liu Y, White CC. et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695
59. Xu JX, Albrecht E, Viergutz T. et al. Perilipin, C/EBPalpha, and C/EBPbeta mRNA abundance in longissimus muscle and different adipose tissues of Holstein and Charolais cattle. Meat Sci. 2009;83:120-126
60. Chen J, Dodson MV, Jiang Z. Cellular and molecular comparison of redifferentiation of intramuscular- and visceral-adipocyte derived progeny cells. Int J Biol Sci. 2010;6:80-88
61. Chen C, Ai H, Ren J. et al. A global view of porcine transcriptome in three tissues from a full-sib pair with extreme phenotypes in growth and fat deposition by paired-end RNA sequencing. BMC Genomics. 2011;12:448
62. Hishikawa D, Hong YH, Roh SG. et al. Identification of genes expressed differentially in subcutaneous and visceral fat of cattle, pig, and mouse. Physiol Genomics. 2005;21:343-350
63. Li M, Wu H, Wang T. et al. Co-methylated Genes in Different Adipose Depots of Pig are Associated with Metabolic, Inflammatory and Immune Processes. Int J Biol Sci. 2012;8:831-837
64. Li M, Wu H, Luo Z. et al. An atlas of DNA methylomes in porcine adipose and muscle tissues. Nat Commun. 2012;3:850
65. Yim JE, Heshka S, Albu J. et al. Intermuscular adipose tissue rivals visceral adipose tissue in independent associations with cardiovascular risk. Int J Obes (Lond). 2007;31:1400-1405
66. Bong JJ, Cho KK, Baik M. Comparison of gene expression profiling between bovine subcutaneous and intramuscular adipose tissues by serial analysis of gene expression. Cell Biol Int. 2010;34:125-133
67. Albrecht E, Gotoh T, Ebara F. et al. Cellular conditions for intramuscular fat deposition in Japanese Black and Holstein steers. Meat Sci. 2011;89:13-20
68. Albrecht E, Gotoh T, Viergutz T. et al. Gene expression profile of intramuscular and subcutaneous fat in Japanese Black and Holstein steers. Proc 9th World Congr genetics Appl Livest Prod (WCGALP), Leipzig, Germany. 2010 (ISBN 978-3-00-031608-1)
69. Zhou G, Wang S, Wang Z. et al. Global comparison of gene expression profiles between intramuscular and subcutaneous adipocytes of neonatal landrace pig using microarray. Meat Sci. 2010;86:440-450
70. Tchkonia T, Lenburg M, Thomou T. et al. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. Am J Physiol Endocrinol Metab. 2007;292:E298-E307
71. Walden TB, Hansen IR, Timmons JA. et al. Recruited vs. nonrecruited molecular signatures of brown, "brite," and white adipose tissues. Am J Physiol Endocrinol Metab. 2012;302:E19-E31
72. Ortiz-Colon G, Grant AC, Doumit ME. et al. Bovine intramuscular, subcutaneous, and perirenal stromal-vascular cells express similar glucocorticoid receptor isoforms, but exhibit different adipogenic capacity. J Anim Sci. 2009;87:1913-1920
73. Gotoh T, Albrecht E, Teuscher F. et al. Differences in muscle and fat accretion in Japanese Black and European cattle. Meat Sci. 2009;82:300-308
74. Dodson MV, Hausman GJ, Guan L. et al. Lipid metabolism, adipocyte depot physiology and utilization of meat animals as experimental models for metabolic research. Int J Biol Sci. 2010;6:691-699
75. Albrecht E, Komolka K, Kudzinski J. et al. Agouti Revisited: Transcript Quantification of the ASIP Gene in Bovine Tissues Related to Protein Expression and Localization. PLoS One. 2012;7:e35282
76. Lehr S, Hartwig S, Lamers D. et al. Identification and validation of novel adipokines released from primary human adipocytes. Mol Cell Proteomics. 2012;11:M111
77. Sarr O, Louveau I, Le Huerou-Luron I. et al. Adipose tissue proteomes of intrauterine growth-restricted piglets artificially reared on a high-protein neonatal formula. J Nutr Biochem. 2012;23:1417-1424
78. Gondret F, Guevel B, Com E. et al. A comparison of subcutaneous adipose tissue proteomes in juvenile piglets with a contrasted adiposity underscored similarities with human obesity. J Proteomics. 2012;75:949-961
79. Gondret F, Guitton N, Guillerm-Regost C. et al. Regional differences in porcine adipocytes isolated from skeletal muscle and adipose tissues as identified by a proteomic approach. J Anim Sci. 2008;86:2115-2125
80. Perez-Perez R, Ortega-Delgado FJ, Garcia-Santos E. et al. Differential proteomics of omental and subcutaneous adipose tissue reflects their unalike biochemical and metabolic properties. J Proteome Res. 2009;8:1682-1693
81. Peinado JR, Jimenez-Gomez Y, Pulido MR. et al. The stromal-vascular fraction of adipose tissue contributes to major differences between subcutaneous and visceral fat depots. Proteomics. 2010;10:3356-3366
82. Schicher M, Morak M, Birner-Gruenberger R. et al. Functional proteomic analysis of lipases and esterases in cultured human adipocytes. J Proteome Res. 2010;9:6334-6344
83. Rajesh RV, Heo GN, Park MR. et al. Proteomic analysis of bovine omental, subcutaneous and intramuscular preadipocytes during in vitro adipogenic differentiation. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:234-244
84. Roca-Rivada A, Alonso J, Al-Massadi O. et al. Secretome analysis of rat adipose tissues shows location-specific roles for each depot type. J Proteomics. 2011;74:1068-1079
85. Trayhurn P, Drevon CA, Eckel J. Secreted proteins from adipose tissue and skeletal muscle - adipokines, myokines and adipose/muscle cross-talk. Arch Physiol Biochem. 2011;117:47-56
86. Trayhurn P, Wood IS. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br J Nutr. 2004;92:347-355
87. Alvarez-Llamas G, Szalowska E, de Vries MP. et al. Characterization of the human visceral adipose tissue secretome. Mol Cell Proteomics. 2007;6:589-600
88. Zvonic S, Lefevre M, Kilroy G. et al. Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol Cell Proteomics. 2007;6:18-28
89. Kim J, Choi YS, Lim S. et al. Comparative analysis of the secretory proteome of human adipose stromal vascular fraction cells during adipogenesis. Proteomics. 2010;10:394-405
90. Rosenow A, Arrey TN, Bouwman FG. et al. Identification of novel human adipocyte secreted proteins by using SGBS cells. J Proteome Res. 2010;9:5389-5401
91. Zhong J, Krawczyk SA, Chaerkady R. et al. Temporal profiling of the secretome during adipogenesis in humans. J Proteome Res. 2010;9:5228-5238
92. Dahlman I, Elsen M, Tennagels N. et al. Functional annotation of the human fat cell secretome. Arch Physiol Biochem. 2012;118:84-91
93. Taube A, Lambernd S, van Echten-Deckert G. et al. Adipokines promote lipotoxicity in human skeletal muscle cells. Arch Physiol Biochem. 2012;118:92-101
94. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83-90
95. McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci U S A. 1997;94:12457-12461
96. Febbraio MA, Pedersen BK. Contraction-induced myokine production and release: is skeletal muscle an endocrine organ? Exerc Sport Sci Rev. 2005;33:114-119
97. Pedersen BK. Edward F. Adolph distinguished lecture: muscle as an endocrine organ: IL-6 and other myokines. J Appl Physiol. 2009;107:1006-1014
98. Steensberg A, Keller C, Starkie RL. et al. IL-6 and TNF-alpha expression in, and release from, contracting human skeletal muscle. Am J Physiol Endocrinol Metab. 2002;283:E1272-E1278
99. Pedersen BK, Steensberg A, Fischer C. et al. Searching for the exercise factor: is IL-6 a candidate? J Muscle Res Cell Motil. 2003;24:113-119
100. Pedersen BK. Muscles and their myokines. J Exp Biol. 2011;214:337-346
101. Broholm C, Laye MJ, Brandt C. et al. LIF is a contraction-induced myokine stimulating human myocyte proliferation. J Appl Physiol. 2011;111:251-259
102. Hojman P, Dethlefsen C, Brandt C. et al. Exercise-induced muscle-derived cytokines inhibit mammary cancer cell growth. Am J Physiol Endocrinol Metab. 2011;301:E504-E510
103. Pedersen L, Olsen CH, Pedersen BK. et al. Muscle-derived expression of the chemokine CXCL1 attenuates diet-induced obesity and improves fatty acid oxidation in the muscle. Am J Physiol Endocrinol Metab. 2012;302:E831-E840
104. Seldin MM, Peterson JM, Byerly MS. et al. Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J Biol Chem. 2012;287:11968-11980
105. Lambernd S, Taube A, Schober A. et al. Contractile activity of human skeletal muscle cells prevents insulin resistance by inhibiting pro-inflammatory signalling pathways. Diabetologia. 2012;55:1128-1139
106. Nikolic N, Skaret BS, Tranheim KE. et al. Electrical pulse stimulation of cultured human skeletal muscle cells as an in vitro model of exercise. PLoS One. 2012;7:e33203
107. Henningsen J, Rigbolt KT, Blagoev B. et al. Dynamics of the skeletal muscle secretome during myoblast differentiation. Mol Cell Proteomics. 2010;9:2482-2496
108. Henningsen J, Pedersen BK, Kratchmarova I. Quantitative analysis of the secretion of the MCP family of chemokines by muscle cells. Mol Biosyst. 2011;7:311-321
109. Bostrom P, Wu J, Jedrychowski MP. et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463-468
110. Mormeneo E, Jimenez-Mallebrera C, Palomer X. et al. PGC-1alpha induces mitochondrial and myokine transcriptional programs and lipid droplet and glycogen accumulation in cultured human skeletal muscle cells. PLoS One. 2012;7:e29985
111. Kiefer FW, Vernochet C, O'Brien P. et al. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med. 2012;18:918-925
112. Koncarevic A, Kajimura S, Cornwall-Brady M. et al. A Novel Therapeutic Approach to Treating Obesity through Modulation of TGFbeta Signaling. Endocrinology. 2012;153:3133-3146
113. Kleiner S, Mepani RJ, Laznik D. et al. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proc Natl Acad Sci U S A. 2012;109:9635-9640
114. Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257-262
115. Lin J, Wu H, Tarr PT. et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797-801
116. Seale P, Kajimura S, Yang W. et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38-54
117. Seale P, Bjork B, Yang W. et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961-967
118. Kajimura S, Seale P, Kubota K. et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154-1158
119. Seale P, Conroe HM, Estall J. et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96-105
120. Tseng YH, Kokkotou E, Schulz TJ. et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000-1004
121. Sun L, Xie H, Mori MA. et al. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958-965
122. Lametsch R, Bendixen E. Proteome analysis applied to meat science: characterizing postmortem changes in porcine muscle. J Agric Food Chem. 2001;49:4531-4537
123. Bouley J, Chambon C, Picard B. Mapping of bovine skeletal muscle proteins using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2004;4:1811-1824
124. Jia X, Hollung K, Therkildsen M. et al. Proteome analysis of early post-mortem changes in two bovine muscle types: M. longissimus dorsi and M. semitendinosis. Proteomics. 2006;6:936-944
125. Laville E, Sayd T, Terlouw C. et al. Comparison of sarcoplasmic proteomes between two groups of pig muscles selected for shear force of cooked meat. J Agric Food Chem. 2007;55:5834-5841
126. Kim NK, Joh JH, Park HR. et al. Differential expression profiling of the proteomes and their mRNAs in porcine white and red skeletal muscles. Proteomics. 2004;4:3422-3428
127. Lametsch R, Kristensen L, Larsen MR. et al. Changes in the muscle proteome after compensatory growth in pigs. J Anim Sci. 2006;84:918-924
128. Bouley J, Meunier B, Chambon C. et al. Proteomic analysis of bovine skeletal muscle hypertrophy. Proteomics. 2005;5:490-500
129. Wang J, Chen L, Li D. et al. Intrauterine growth restriction affects the proteomes of the small intestine, liver, and skeletal muscle in newborn pigs. J Nutr. 2008;138:60-66
130. Xu Y, Qian H, Feng X. et al. Differential proteome and transcriptome analysis of porcine skeletal muscle during development. J Proteomics. 2012;75:2093-2108
131. Schennink A, Bovenhuis H, Leon-Kloosterziel KM. et al. Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milk-fat composition. Anim Genet. 2009;40:909-916
132. Ryu J, Kim Y, Kim C. et al. Association of bovine carcass phenotypes with genes in an adaptive thermogenesis pathway. Mol Biol Rep. 2012;39:1441-1445
133. Khatib H, Zaitoun I, Wiebelhaus-Finger J. et al. The association of bovine PPARGC1A and OPN genes with milk composition in two independent Holstein cattle populations. J Dairy Sci. 2007;90:2966-2970
134. White SN, Casas E, Allan MF. et al. Evaluation in beef cattle of six deoxyribonucleic acid markers developed for dairy traits reveals an osteopontin polymorphism associated with postweaning growth. J Anim Sci. 2007;85:1-10
135. Kim JM, Lee KT, Lim KS. et al. Effects of p.C430S polymorphism in the PPARGC1A gene on muscle fibre type composition and meat quality in Yorkshire pigs. Anim Genet. 2010;41:642-645
136. Gandolfi G, Cinar MU, Ponsuksili S. et al. Association of PPARGC1A and CAPNS1 gene polymorphisms and expression with meat quality traits in pigs. Meat Sci. 2011;89:478-485
137. Kim JM, Lim KS, Lee EA. et al. Polymorphisms of the 5' regulatory region of the porcine PPARGC1A gene and the effects on muscle fiber characteristics and meat quality. Mol Biol Rep. 2012;39:3933-3942
138. Erkens T, Bilek K, Van ZA. et al. Two new splice variants in porcine PPARGC1A. BMC Res Notes. 2008;1:138
139. Wang J, Wang C, Tian R. et al. Sequence variants in the bovine PRDM16 gene associated with body weight in Chinese cattle breeds. Genet Mol Res. 2012;11:746-755
140. Levin K, Daa SH, Alford FP. et al. Morphometric documentation of abnormal intramyocellular fat storage and reduced glycogen in obese patients with Type II diabetes. Diabetologia. 2001;44:824-833
141. Corpeleijn E, Hessvik NP, Bakke SS. et al. Oxidation of intramyocellular lipids is dependent on mitochondrial function and the availability of extracellular fatty acids. Am J Physiol Endocrinol Metab. 2010;299:E14-E22
142. Goodpaster BH, He J, Watkins S. et al. Skeletal muscle lipid content and insulin resistance: evidence for a paradox in endurance-trained athletes. J Clin Endocrinol Metab. 2001;86:5755-5761
143. Schrauwen-Hinderling VB, Hesselink MK, Schrauwen P. et al. Intramyocellular lipid content in human skeletal muscle. Obesity (Silver Spring). 2006;14:357-367
144. Meex RC, Schrauwen P, Hesselink MK. Modulation of myocellular fat stores: lipid droplet dynamics in health and disease. Am J Physiol Regul Integr Comp Physiol. 2009;297:R913-R924
145. Zhang H, Wang Y, Li J. et al. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. J Proteome Res. 2011;10:4757-4768
146. Gallagher D, Kuznia P, Heshka S. et al. Adipose tissue in muscle: a novel depot similar in size to visceral adipose tissue. Am J Clin Nutr. 2005;81:903-910
147. Vettor R, Milan G, Franzin C. et al. The origin of intermuscular adipose tissue and its pathophysiological implications. Am J Physiol Endocrinol Metab. 2009;297:E987-E998
148. Aguiari P, Leo S, Zavan B. et al. High glucose induces adipogenic differentiation of muscle-derived stem cells. Proc Natl Acad Sci U S A. 2008;105:1226-1231
149. Scheele C, Nielsen S, Kelly M. et al. Satellite Cells Derived from Obese Humans with Type 2 Diabetes and Differentiated into Myocytes In Vitro Exhibit Abnormal Response to IL-6. PLoS One. 2012;7:e39657
150. Karst S, Cheng R, Schmitt AO. et al. Genetic determinants for intramuscular fat content and water-holding capacity in mice selected for high muscle mass. Mamm Genome. 2011;22:530-543
151. Joe AW, Yi L, Even Y. et al. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells. 2009;27:2563-2570
152. Joe AW, Yi L, Natarajan A. et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153-163
153. Majka SM, Fox KE, Psilas JC. et al. De novo generation of white adipocytes from the myeloid lineage via mesenchymal intermediates is age, adipose depot, and gender specific. Proc Natl Acad Sci U S A. 2010;107:14781-14786
154. Majka SM, Barak Y, Klemm DJ. Concise review: adipocyte origins: weighing the possibilities. Stem Cells. 2011;29:1034-1040
155. Albrecht E, Gotoh T, Ebara F. et al. Technical note: Determination of cell-specific gene expression in bovine skeletal muscle tissue using laser microdissection and reverse-transcription quantitative polymerase chain reaction. J Anim Sci. 2011;89:4339-4343
156. Kouba M, Sellier P. A review of the factors influencing the development of intermuscular adipose tissue in the growing pig. Meat Sci. 2011;88:213-220
Author contact
![]() Corresponding author: Dr. S. Maak, Research Unit Muscle Biology and Growth, Leibniz-Institute for Farm Animal Biology (FBN), W.-Stahl-Allee 2, D-18196 Dummerstorf, Germany; Tel: +49-38280-68850; FAX: +49-38208-68852; E-mail: maakde.
Corresponding author: Dr. S. Maak, Research Unit Muscle Biology and Growth, Leibniz-Institute for Farm Animal Biology (FBN), W.-Stahl-Allee 2, D-18196 Dummerstorf, Germany; Tel: +49-38280-68850; FAX: +49-38208-68852; E-mail: maakde.

 Global reach, higher impact
Global reach, higher impact