ISSN: 1839-9940Journal of Genomics
J Genomics 2014; 2:104-117. doi:10.7150/jgen.8178 This volume Cite
Review
Meiotic Sex Chromosome Inactivation in Drosophila
Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, São Paulo, Brazil 05508.
Abstract
In several different taxa, there is indubitable evidence of transcriptional silencing of the X and Y chromosomes in male meiotic cells of spermatogenesis. However, the so called meiotic sex chromosome inactivation (MSCI) has been recently a hot bed for debate in Drosophila melanogaster. This review covers cytological and genetic observations, data from transgenic constructs with testis-specific promoters, global expression profiles obtained from mutant, wild-type, larvae and adult testes as well as from cells of different stages of spermatogenesis. There is no dispute on that D. melanogaster spermatogenesis presents a down-regulation of X chromosome that does not result from the lack of dosage compensation. However, the issue is currently focused on the level of reduction of X-linked expression, the precise time it occurs and how many genes are affected. The deep examination of data and experiments in this review exposes the limitations intrinsic to the methods of studying MSCI in D. melanogaster. The current methods do not allow us to affirm anything else than the X chromosome down-regulation in meiosis (MSCI). Therefore, conclusion about level, degree or precise timing is inadequate until new approaches are implemented to know the details of MSCI or other processes involved for D. melanogaster model.
Keywords: down-regulation, X chromosome, spermatogenesis, testis, male germline.
Introduction
Meiotic sex chromosome inactivation (MSCI) is the early transcriptional silencing of the X and Y chromosomes that occurs in the male germline meiotic phase [1-3]. MSCI has been observed in a vast range of taxonomic groups such as mammals, nematodes and grasshoppers [4-6]. In mice, the organism where MSCI has been best characterized, cytological evidence shows that the X and Y chromosomes are compartmentalized in a nuclear subdomain of pachytene cells [4,7]. At this stage, extensive chromatin modifications occur on the sex-chromosome that results in significant down-regulation of the X-linked expression [8-10].
Several hypotheses have been proposed for the evolution of MSCI. In one hypothesis, MSCI could have evolved to prevent detrimental products from the recombination of non-homologous sex-chromosomes such as those generate in ectopic changes and double strand breaks [3]. Recently, studies have shown that MSCI is a special example of a general mechanism in which unpaired chromosomes are silenced [11-13]. The general phenomenon, called meiotic silencing of unsynapsed chromatin (MSUC), has been observed in mouse and worms and may avoid the spread of aneuploidy [11-13]. However, there is no recombination by crossing over in D. melanogaster male meiosis [14] and therefore no need to evolve special mechanisms to avoid harmful recombination of unsynapsed chromosomes [3]. Therefore, for several years, it has been thought that MSCI probably evolved in the Drosophila genus for other reasons [3]. However, two recently published data revived the idea that MSCI in Drosophila is connect to the evolution of mechanisms that avoid harmful recombination. First, not all species in the genus lack male recombination, as crossing over does occur in males of D. ananassae from natural populations [15]. Second, it is possible that the dot chromosome of D. melanogaster was ancestrally a differentiated X chromosome that still bears sequence and architectural features used in sex chromosome-specific regulatory mechanisms, including in meiosis [16]. Similar, if the chiasmatic status has been lost in D. melanogaster, it is possible that meiotic down-regulation remains among X-linked genes [3].
Another hypothesis for the evolution of MSCI, “the drive hypothesis”, suggests that the meiotic silencing of the X chromosome is advantageous because it prevents meiotic expression of sex-ratio distorters which are observed in different taxa but are very frequently found in the Drosophila genus [17]. Finally, the sexual antagonism and X inactivation (SAXI) hypothesis proposes that MSCI is driven by sexual antagonistic effects of X-linked genes [18]. X chromosomes are expected to be feminized, in terms of gene content, because they spend more evolutionary time in females than in males and should therefore accumulate genes which are beneficial for females [18, 19]. If female-beneficial genes are detrimental to males, it may be advantageous to reduce or eliminate their expression in the late stages of spermatogenesis [18].
MSCI may explain several puzzling observations made in recent genome evolution studies. In Drosophila and mammals, there is an excess of new genes on the autosomes which were formed by duplication of X-linked genes [20-24]. These new genes often acquire male expression and male-related functions [20-24]. In addition, the X chromosome is generally depleted of genes expressed more highly in testis than ovary, that is testis-biased genes [25-27]. MSCI, among other hypotheses, has been proposed as explanation for the evolution of both observations as testis-expressed genes located in the autosomes would still be expressed during the inactivation of the X chromosome [1,21]. Indeed, such compensatory meiotic expression has been found for autosomal genes formed by RNA-based duplication of X-linked genes (retrogenes) in mammals and Drosophila [28, 29].
The existence of MSCI in D. melanogaster was proposed in the 1970s based on observations of major male sterility in flies with X - autosome translocations [1]. Advances from genomics and transcriptomics coupled with new cell biological techniques have allowed scientists to initiate and continue investigations into the existence and evolutionary impact of MSCI in many taxa, including D. melanogaster [e.g., 28-32]. However, recent conflicting results have placed MSCI in Drosophila at the center of a debate [29-31, 33-39]. Although there is evidence of down-regulation of the X chromosome, the magnitude and the exact timing of its occurrence during spermatogenesis put in doubt if the phenomenon is as strong in D. melanogaster as it is in mammals or if X chromosome regulation is governed by another, previously undescribed mechanism. Here I revisit all current available data in favor and against the existence of MSCI in D. melanogaster in order to evaluate these data related to the debate and stimulate projects and ideas for its resolution.
Genetic and Cytological Observations of MSCI in Drosophila
In 1972, Lifschytz and Lindsley [1] published a seminal work in which they assembled genetic and cytological observations to propose the hypothesis that X inactivation is a universal requirement for proper spermatogenesis in all male-heterogametic organisms. They based this conclusion on the genetic observations that D. melanogaster males with reciprocal translocations between X chromosomes and autosomes were often sterile whereas males with translocations between autosomes are generally fertile [1]. This sterility phenotype is dominant as fertility was not restored by the addition of segments including the translocation breakpoints and their neighboring genes [1]. This observation suggested that the translocations affected the fly's biology at the chromosomal level and that the X chromosome and the autosomes should be asynchronously controlled in order for normal spermatogenesis to occur [1]. Although translocations between the X chromosome and chromosome 2 or 3 cause sterility, translocations between the X and chromosome 4 are male fertile [1]. These observations led Lifschytz and Lindsley [1] to suggest that chromosomes X and 4 activities are relatively in phase during spermatogenesis four decades ago, before the discovery that it is possible that the dot chromosome was ancestrally a differentiated X chromosome [16].
Lifschytz and Lindsley's [1] cytological evidence for the importance of MSCI came from Cooper's [40] and from their own observations of the meiotic condensation of the sex chromosomes before the autosomes during normal D. pseudoobscura spermatogenesis and in D. melanogaster X/0 males [1]. However, even Cooper [40] noted that his description “should be regarded as wholly tentative” first because his work only synthesized and reinterpreted others' work and second because of the prominent difficulties in doing cytological studies in D. melanogaster meiotic phase. Cytological evidence of MSCI in Drosophila has been disputed since then. Although all studies agree that sex chromosome condensation begins earlier than the condensation of autosomes in spermatocytes (the cell type in which meiosis occurs), there are periods during meiosis that the X chromosome is decondensed and other periods in which no differential condensation between chromosomes is recognized [1,3, 40,41].
Indeed, McKee and Handel [3] specifically claimed that there is no MSCI in Drosophila based on the arguments that i) the cytological evidence was inconclusive [1, 3, 40, 41] and ii) D. melanogaster male meiosis is achiasmatic, i.e. does not involve crossing over [14]. The authors had proposed the hypothesis that MSCI evolved in different taxa to prevent physical DNA damage originating from meiotic recombination of non-homologous sex-chromosomes [3]. The connection between the lack of male meiotic recombination and supposed lack of MSCI in D. melanogaster would fit the predictions of their hypothesis [3]. However, today it is known that lack of meiotic recombination in males is not extended to the entire genus [15].
The differential distribution of histone acetylation on the X chromosome and autosomes provides another type of cytological indirect support for MSCI in Drosophila [42]. If X inactivation occurs, the X chromatin may be enriched with histone H4 underacetylated at all residues except lysine 12 (H4Ac12) which are normally found in heterochromatin of somatic cells. Analyses of those histones in late spermatocytes showed that they are not labeling autosomes, but might be labeling the X-Y cluster in Figure 6B from [43].
Surprisingly, despite all contrasting and disputing data, it has been several years since anyone has published cytological data on MSCI. It might be that Drosophila male germline staining and chromosomal topology measurement were not feasible [40]. However, new advances in visualization techniques, if applied to the study of MSCI, may resolve this issue.
Expression Analyses of Transgenic Constructs
In the absence of continued cytological studies of MSCI, several groups have used genetic evidence to assess its presence and impacts on Drosophila genome evolution. The transcriptional silencing expected by meiotic sex chromosomal inactivation can be observed also by the lack of or significant reduction of expression of X-linked genes in male meiosis. Transgenic constructions combining reporter genes with a testis-specific promoter have been proven to be quite useful in testing the MSCI hypothesis [30,38,39,44]. These studies are based on the assumption that if the X chromosome is inactivated during male meiosis, reporter gene expression levels will be lower when the reporters are inserted in the X chromosome compared to those inserted in the autosomes (Figure 1) [30,44]. First support to the MSCI hypothesis using this approach came from an early study in which the expression of constructs with a modified form of β-tubulin driven by a testis-specific promoter were consistent with the early cessation of X chromosomal expression in D. melanogaster spermatogenesis [44]. Analysis of protein abundance in electrophoresis gels to estimate gene expression revealed that X-linked insertions produced much less protein than autosomal ones, as expected by the X inactivation hypothesis [44].
Recently, the expression profile of a reporter gene driven by the promoter of the testis-specific ocnus (ocn) gene [45] provided a more rigorous experimental test for MSCI [30]. In contrast to earlier β-tubulin experiments [44], this study measured both transcript and protein abundances for a large number of independent insertions, allowing for statistical analysis of the effects, or lack of effects, of MSCI in D. melanogaster. In addition, the transgene was also inserted within a sequence that shields the impact of external transcription regulators avoiding any effect from repressors that could be preferentially bound to the X chromosome [30].
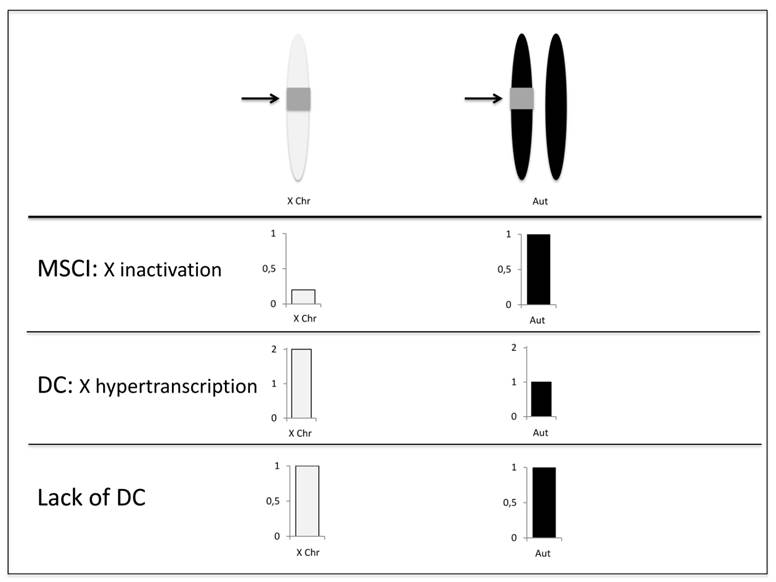
A major advantage of the construct insertion approach is that the results are conservative with respect to dosage compensations effects (Figure 1). Dosage compensation in Drosophila is achieved by hypertranscription of the X chromosome in males [46]. Therefore, the reduced expression found for X-linked insertions could be a result of the lack of dosage compensation. However, autosomal insertions were made heterozygous, i.e. in one copy [30]. Therefore, if the X chromosome is not hypertranscribed, expressions of single copy autosomal and X-linked insertions are expected to be equal. Alternatively, any level of X chromosome hypertranscription would lead to a higher expression of the single copy X-linked insertions. None of the outcomes involving dosage compensation explain the observed results - lower expression of transgenes inserted in the X chromosome - which are consistent with X inactivation [30].
Some drawbacks of the transgenic experimental approach have been pointed out, including the fact that the observations were restricted to a single promoter and the transgene insertions do not cover the entire X chromosome [29,34]. However, using modifications of the ocn transgenic approach the same group has refuted those shortcomings one by one [38, 39]. First, a fine-scale map of the X inactivation in the male germline was generated by the insertion of 112 independent insertions of the same construct [38]. Finding consistent down-regulation of widespread X-linked insertions excluded the possibility that some large chromosomal neighborhoods of the X chromosome escape inactivation [38]. Second, three other promoters of X-linked testis-expressed genes were analyzed [39]. The experiments not only prove that the germline down-regulation of X-linked insertions is pertinent to promoters other than ocn, but also shows that the increased expression associated to the relocation to autosomes occurs even for genes that are originally located on the X chromosome and therefore adapted to its local chromosomal environment [39].
In addition, the same study further tested the possibility that the reduction of X-linked expression arises from an intrinsic property of their transformations by profiling the expression of a somatic transgene, mini-white, with expression driven by testis-specific promoters [39]. X-linked mini-white insertions were found to be more highly expressed in males than autosomal heterozygous insertions, consistent with patterns of dosage compensation (Figure 1).
It is important to note that although expression level and protein abundance measurements were done with the entire testis, the general down-regulation observed from the X-linked insertions should be applied to the meiotic and post-meiotic phases in males instead of the whole spermatogenesis process [30,39]. Most of the report genes are not expressed or are expressed in a very low level in the mitotic phase of testis as shown in the in situ hybridizations excluding the effects of mitotic regulation of the X chromosome [30,39]. Taken together, all those careful experiments using transgenic construct provide compelling evidence in favor of the MSCI hypothesis.
Detecting MSCI using transgenic constructs and random insertions. Autosomal insertions were made heterozygous in order to disentangle the effects of MSCI and dosage compensation [30]. As only one homolog of the autosome contains the insertion, if there is MSCI, the X-linked inserts will show relatively lower expression. In the case of lack of dosage compensation, X-linked and autosomal inserts will not show differential expression. For any level of dosage compensation (hypertranscription), the X-linked insertions will show higher expression than autosomal ones as the latter is presented in only one copy. The arrows point for the insertions (gray squares). Expression of X-linked and autosomal insertions are shown in white and black respectively.
Global Expression Analyses of Spermatogenesis
MSCI has been also investigated by microarray global gene expression analysis of spermatogenesis [29]. D. melanogaster spermatogenesis occurs in developmental stages - mitotic, meiotic and pos-meiotic - that are progressively distributed within an adult testis [40,47]. Apical regions contain the first gonial stem cells and subsequently produced mitotic cells while distal regions contain the packed nuclear sperm heads [47]. A detailed schematic diagram describing the different phases of Drosophila spermatogenesis and how they are arrayed in the Drosophila testis can be found in Figure 2 in [47].
The dissection of three distinct regions of the testis which were respectively enriched with mitotic, meiotic and post-meiotic cells composed the first spermatogenic-stage-specific transcriptome of D. melanogaster [29]. Comparison of X chromosome and autosome expression under a Bayesian framework revealed a significant, although small (around 10%), reduction of X-linked gene expression in meiosis, in agreement with signal of MSCI [29]. The same study also evaluated the evolutionary consequences of X down-regulation by correlating it with the frequent relocation of retrogenes to the autosomes [29], as observed previously in [21, 24].
However, Meiklejohn et al. [33] revisited the data of the stage-specific spermatogenic transcriptome [29]; with additional experiments, they suggested contrary conclusions about MSCI. Briefly, they found no evidence for X-linked genes expression reduction in meiotic cells, whereas mitotic germline cells seem to lack dosage compensation [33]. They also analyzed the expression of testis-specific ocnus transgenes [30] finding lower expression of X-linked inserts in both mitotic and meiotic cells [33]. Since transgenic insertions are heterozygous and therefore their expression reduction is incompatible with lack of dosage compensation, the authors hypothesized that another unrecognized mechanism regulates X chromosome expression during spermatogenesis [33]. These conflicting results raised considerable debate over MSCI in D. melanogaster [33, 34, 36; for criticism, see 29 and 35]. In the following, I describe the different major opinions and discuss how these arguments were based on a lack of understanding of current experimental limitations and even on erroneous interpretation of previous experimental data for the study of MSCI.
Mitotic Purity
Meiklejohn et al. [33] major point against MSCI in Drosophila is that mitotic samples dissected in the stage-specific spermatogenic transcriptome [29] already show lower expression levels of X-linked genes when compared to autosomal ones. If levels of expression between autosomes and X chromosome are not equal so early in spermatogenesis, the pattern should not be a result of MSCI. However, Meiklejohn et al. [33] did not take into account the level of mitotic purity in mitotic samples, i.e. the level of contamination with meiotic cells in this sample.
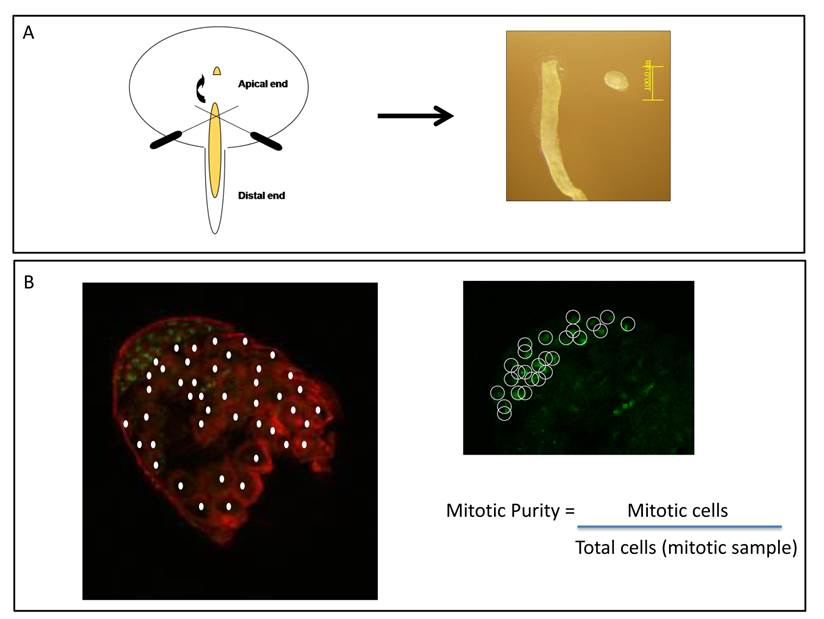
In order to test X inactivation by comparing the transcriptome of different spermatogenic stages it is important to understand D. melanogaster testis cell biology and the basis of cell dissections. Due to coiled form of testes and how cells at different spermatogenic stages are distributed and located inside it, the full separation of mitotic and meiotic cells is unlikely. From Figure 1 in Vibranovski et al. [29], partially reproduced and adapted here in Figure 2B, one can easily identify that the mitotic sample is not pure; i.e. does not contain just mitotic cells. Based on the differential DNA staining for diploid and haploid cells (Figure 2B), mitotic samples clearly contain a significant number of meiotic cells [29]. Mitotic purity, defined as the percentage of mitotic cells in the mitotic sample, is approximately 30%. Note that there is always an experimental variation within the apical tip size excised from the whole testis.
Dissection of three distinct testis regions gave samples that were enriched, not specified, with a certain cell type [29]. Due to the limit of technology available, the intention in this experiment was to increase the proportion of, for instance, the mitotic cells in the sample when compared to the whole testis. Such a procedure does not imply that the mitotic cell type would be the only type, or even the major type in the sample [29]. Factually neglecting such limitation has had a negative impact for drawing an incorrect conclusion about MSCI in [33]. On one hand, if no difference between mitotic and meiotic X chromosome expression is observed, concluding that there is no MSCI in D. melanogaster is precipitous as the presence of meiotic cells in mitotic sample likely diffuse the MSCI signal. On the other hand, when detecting statistically significant down-regulation of the X chromosome as in [29], it should be taken as evidence of MSCI but no further interpretation whether MSCI is global, relatively weak or not severe should be reached, because of the very property of the data created by available techniques.
Mitotic purity. Percentage of mitotic cells contained in D. melanogaster apical tips, the mitotic sample. A. Scheme of wild-type testis being dissected in a PBS drop. Left panel: apical tip is cut off from testes using 0.25mm diameter insect pins. Right panel: picture of an apical tip detached from the remaining testis. B. Adapted from Figure 1 in [29]: apical tip with indirect immunofluorescence staining alpha-tubulin and DNA in red and green, respectively. Left panel: meiotic cells shown by the filled circles are identified as those greater in size and contained less bright DNA staining correspond to haploid cells. Right panel: the same apical tip shown on the left for green immunofluorescence panel only. Mitotic cells shown by the open circles were identified as those containing brighter DNA staining, the diploid ones. Mitotic purity was estimated by the ratio between the number of mitotic cells and total number of cells [29].
Mitotic Dosage Compensation
Meiklejohn et al. [33] also took advantage of the spermatogenic-stage-specific transcriptome of D. melanogaster to investigate germline dosage compensation (see more details in Expression Analyses of Mutants of Early Development Arrest). The authors tested whether the X chromosome has the same expression level as the autosomes by comparing the expression of genes linked to the X and to the autosomes in the mitotic sample. According to Meiklejohn et al. [33], the mitotic sample presents a more homogenous cell population than the meiotic cell population and is therefore free from any regulatory phenomena specific to meiosis. Dosage compensation was assessed by the absolute difference in expression between the X chromosome and autosomes in mitotic cells collected from the apical tips (Figure 2B). Autosomal genes showed significantly higher median expression than the X-linked genes, which corresponds to the expression differences expected in the absence of dosage compensation (adapted in Table 1 here) [33].
However, the techniques for detecting the effects of MSCI were designed, based on the specific tissue structure of testis that does not allow complete separation by dissection of cells in different spermatogenesis stages, in a way that the mitotic sample was mixed up with meiotic cells (Figure 2). Therefore, a more likely scenario is that the lower expression of the X chromosome in mitotic sample was a result of MSCI. The difficulty in obtaining pure mitotic cell samples makes the use of the mitotic sample absolute expression to test dosage compensation difficult to interpret [33]. Testing the existence of MSCI by comparing expression between mitotic and meiotic samples, however, is effective as it is based on the relative reduction of chromosomal expression between phases [29]. Therefore, even if mitotic samples already show lower expression of the X chromosomes, the relative reduction in meiosis suggests the existence of MSCI. Note in Table 1 that the expression differences between autosomes and the X chromosome seems to be greater in meiotic than in mitotic samples although statistical analyses should be implemented (see below). Unfortunately, all these technique specifics were factually ignored when Meiklejohn et al [33] developed their analyses and interpreted the data in [29].
Values for expression differences between autosomal and X-linked genes obtained from Figure 1 in [33].
| Sample tested | A - X |
|---|---|
| Male somatic cells (control for DC) | -0.02 |
| SL2 cells msl2-RNAi (control for lack of DC) | 0.59*** |
| Mitotic | 0.56*** |
| Meiotic | 0.68*** |
A-X: difference between chromosomal median of log 2 expression
DC: Dosage compensation
SL2 cells msl2-RNAi: D. melanogaster male-like SL2 cells in which mRNA encoding MSL2 was knocked down by RNA interference (RNAi) [48].
***p<0.001 (Mann-Whitney test).
Statistical Analyses for comparison between mitotic and meiotic expression
Meiklejohn et al. [33] tested the small, but previously found by Vibranovski et al. [29] to be significant, reduction of X-linked genes in meiosis in comparison to mitosis using t-tests with false discovery rate (FDR) to correct for multiple tests followed by contingency table tests. Their tests revealed a modest dearth of upregulated genes but no excess of down-regulated genes on the X chromosome in meiotic samples relative to mitotic samples [33]. Given the level of impurity of mitotic samples, it is not surprising that their approach would fail to detect down-regulation, as opposed to our Bayesian framework that fully account for all uncertainties and noise involved in both data collection and modeling steps [29].
Differences between the two statistical methods are clear-cut. In the t-test approach, the first step is implemented to individually classify all the genes as over, under or equally expressed by comparing the expression values of meiosis against mitosis, where each phase has three replicates [33]. In the second step, Fisher exact tests are used to compare the proportions of genes in each class (under or not under expressed) for X chromosome and autosomes [33]. Only in this latter step would X-inactivation be detected in which X chromosome would present significant more meiotic under-expressed genes than the autosomes. This step counts the number of previously classified cases of under-expressing genes without taking into consideration the levels of down-regulation and the variability among cases. In other words, the t-test approach disregards the additional information on the magnitude of expression differences of genes in the different samples that can be used to detect expression differences in the different stages.
In contrast, the Bayesian approach [29] first derives the empirical distributions of the expression differences between meiosis and mitosis of the genes on the X chromosome and autosomes. Second, those empirical distributions are approximated by a finite mixture of two normal distributions. Third, X chromosome and autosomal distributions are compared. This approach avoids the need for using correction for multiple testing because it tests the differential expression at the same time that compares chromosomes [29]. In addition, as the method does not first classify individual genes as over an under expressed, the analysis incorporates the magnitudes of gene expression differences between samples to further increase power to detect differences. The Bayesian analyses were able to detect a larger proportion of differently expressed genes than the t-test approach: ~85% versus ~65% [29, 33].
Additionally, the two major inconsistences between the two approaches raised by Meiklejohn et al. [33] are the following. First, Meiklejohn et al. [33] mistakenly claimed that the Bayesian approach [29] did not exclude genes that are lowly expressed and therefore not significantly expressed above the background. It is possible that the noise generated would mask the real expression differences between chromosomes [33]. However, Vibranovski at al. [29] have also analyzed genes known to be expressed in testis and therefore are above the background expression. Despite the reduction of probe densities, the main conclusions were not changed [29]. Second, Meiklejohn et al. [33] also mistakenly argued that the t-tests account for the variation between replicates values more properly than the Bayesian model [29] as the latter uses mean values for comparisons. However, variances between replicates were negligible smaller than the mean change across stages (e.g., meiosis-mitosis). In addition, the results obtained from the Bayesian approach are robust when using each one of the replicate measurements separately (unpublished but available upon request). Variation in p-values is an inherent feature of the t-test with small sample size such as three replicates.
Interestingly, additional statistical evidence for MSCI is that even Meiklejohn et al. [33] approach was able to detected significant down-regulation of the dot (the 4th chromosome in D. melanogaster) during meiosis in comparison to mitosis [33]. Today, we know that it is possible that the dot in Drosophila has been shown to be ancestrally an X chromosome and to still bear features common to expression regulation of the X chromosome during the spermatogenesis [16].
Differential Somatic Contamination
One source of bias for the interpretation of Meiklejohn et al. [33] for the lower expression of X-linked genes observed in Vibranovski et al. [29] is the differential somatic contamination between the mitotic and meiotic samples. The mitotic sample was obtained with the testis sheath which is a somatic tissue that undergoes complete dosage compensation, presenting X chromosome hypertranscription [29, 33, 46]. The meiotic sample, however, was extracted without the sheath and therefore contains mostly germline cells, which Meiklejohn et al. [33] claimed lacks dosage compensation. In order to test the hypothesis that the somatic tissue contained in the mitotic sample is inflating the X chromosome expression, Meiklejohn et al. [33] analyzed 12 X-linked genes in samples where the testis sheath was detached from the mitotic cells: Table 3 in [33]. For all those genes, the down-regulation observed in the meiotic sample relative to the mitotic sample with sheath disappears when mitotic without sheath was used [33]. This result indicates that sheath contamination in the mitotic sample is a major factor contributing to the differences between X and autosomal expression observed in the meiotic samples [33].
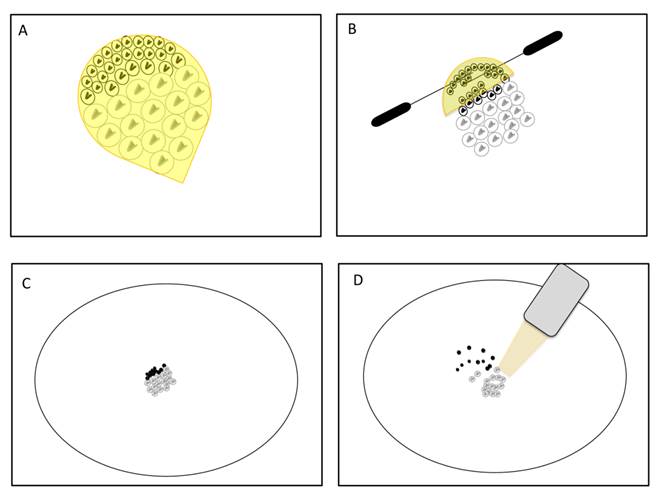
However, this analysis did not take into account that the process of removing testis sheath could significantly decrease the amount of mitotic cells. As mitotic samples also contain meiotic cells (see Mitotic Purity), such a procedure would rather reduce the signal of X inactivation obtained through the comparison between mitotic and meiotic samples. The removal of testis sheaths from the apical tips containing the mitotic cells is not trivial. Similar to the meiotic dissections of Vibranovski et al. [29], mitotic cells can be harvested from the sheath by applying gradual pressure positioning insect pins over the apical tip in an anterior-posterior direction (Figure 3A, B). Several cells, most probably very early spermatogonia, could stick into the upper most region of the sheath tip (Figure 3A, B). In addition, as the dissections are made in PBS buffer drops, the chance for cell diffusion and subsequent loss are higher for smaller cells such as the early mitotic ones (Figure 3C and D). Therefore, until other techniques prove to be more effective, the mitotic without sheath sample is likely to be reduced with mitotic cells causing misinterpretations of the results in [33].
Alternatively, in order to test the possible differential effect of somatic expression, Vibranovski et al [29] analyzed the expression of testis-biased genes, i.e. genes higher expressed in testis than in ovaries. The contribution of somatic expression is reduced in this dataset as it is enriched for genes differentially expressed between germlines. As expected for the MSCI hypothesis, down-regulation of the X-chromosome in meiosis was observed for testis-biased genes [29]. Curiously, all 12 genes analyzed by the mitotic without sheath experiment are not testis-biased genes, expect for the gene CG1835 that clearly show no sheath effect in Table 3 in [33].
Transgene Expression in mitotic and meiotic samples
Finally, Meiklejohn et al. [33] proposed that another unrecognized mechanism regulates X chromosome expression during spermatogenesis. The conclusion was reached by reproducing the analysis of testis-specific ocnus transgene expression [30] but measuring the insertions expression in testis specific stages: the mitotic and the meiotic samples from [29]. Until this analysis, their interpretations of their results were i) that the male germline lacks dosage compensation and ii) MSCI does not exist [33]. However, the transgene experiment showed that X-linked insertions are more lowly expressed in both mitotic and meiotic cells [33]. As in previous transgenic analyses [30], insertions were made heterozygously in autosomes and hemizygously in the X, leading to one copy of each insertion in the genome regardless its location. As dosage compensation is achieved by X hypertranscription [46], expression differences observed between X-linked and autosomal inserts cannot be explained by the absence of dosage compensation (Figure 1). Taken together, the authors suggested that in D. melanogaster there is previously unrecognized mechanism for the down-regulating of the X chromosome expression during spermatogenesis which is lack of neither dosage compensation nor MSCI [33], although no specifics were given to the suspected mechanism and made the hypothesis vague and untestable.
Scheme of technique for testis sheath removal. A. D. melanogaster apical tip containing the mitotic sample surrounded by testis sheath (yellow). Mitotic cells are smaller, show bright DNA staining and are located in the upper most region of the tip. Meiotic cells are larger, show less DNA staining and occupy lower regions. B. Squeezing of cells by insect pins produces gradual pressure over the apical tip in an anterior-posterior direction. C. Groups of cells just after extraction from the apical tip in a PBS drop. D. Cell diffusion before pipetting suction. Note that in both techniques (B and D) mitotic cells have less chance to be sampled.
However, Meiklejohn et al. [33] did not take into account the expression distribution of the reporter genes along the testes showed by in situ experiments [30, 39]. There is almost no expression in mitotic phases for most of the reporter genes as opposed to high levels of expression in the later phases of spermatogenesis including the meiosis [30, 39] which according to [39] “suggests contamination between stages may be an important confounding factor in studies that compare expression between dissected regions of the testes”. Also, the region corresponding to the mitotic cells with no reporter expression [30, 39] is much smaller than the apical tip dissected in [29] and by Meiklejohn et al. [33]. Therefore, lower expression of X-linked inserts observed for both mitotic and meiotic sample is probably due to MSCI effect in meiotic cells present in the mitotic sample [39].
Therefore, given the evidence that i) down regulation of X-linked genes is observed in multiple studies and using multiple methods [29,30,35]; ii) down regulation is observed related to the meiotic stage of spermatogenesis [29,30,35] and iii) mitotic samples are not pure, it appears unlikely that the conclusions of Meiklejohn et al. [33] are the parsimonious solutions. It is more likely to be an erroneous interpretation of the previous published data due to failure to realize that the mitotic samples contain cells from mitotic and meiotic stages of spermatogenesis [29].
Expression Analyses of Mutants of Early Developmental Arrest
Despite all the debate about meiotic sex chromosome inactivation (MSCI) in Drosophila, a lot of discussion regarding the status of X chromosome dosage compensation (DC) in the D. melanogaster male germline has recently intensified in the literature [31, 37, 49]. However, only a few studies have tried to assess and disentangle the contribution of both mechanisms to the evolution of gene expression in the germline [31, 37]. In theory, MSCI is the inactivation of the X chromosome and lack of DC is the absence of hypertranscription of the X chromosome which in males are present in only one copy. However, both phenomena could be seen as lowering the total level of X-linked gene expression in comparison to the total level of autosomal expression. The magnitude of the down-regulation would depend on the power to detect such lower transcription, on the level and incidence of MSCI and/or on the incidence of partial DC. Below, I review the current data examining simultaneously MSCI and DC aspects.
Significant average lower expression of X-linked genes relative to autosomal genes in independent comparisons using whole testis of wild-type flies has been claimed as evidence for lack of DC in D. melanogaster [33]. Specifically, as the magnitude of differential expression of genes linked to X chromosome and autosomes in the germline is similar to what is found for somatic systems lacking dosage compensation [33, 48]. However, whole testis contains all development stages of spermatogenesis. The majority of testis expression comes from spermatocytes (cells of the meiotic phase) [29] and therefore MSCI cannot be ruled out as a contributing factor accounting for the expression differences between genes located in the X chromosome and the autosomes found in whole testis. Moreover, exclusion of MSCI based on apical tips dissections [33] is inherently inaccurate and therefore cannot be used to distinguish between these two possibilities.
However, both DC and MSCI have been investigated through the expression profile of D. melanogaster mutants for spermatogenesis early developmental arrest such as bag of marbles (bam) and benign gonial cell neoplasm (bgcn) [31, 33, 37, 49]. In those mutants, spermatogenesis is disrupted at an early stage and their testes display disproportionate number of cells of the same size as the primary spermatocytes which never undergo subsequent meiotic morphological changes [50]. Comparison between X-linked and autosomal expression in testis of wild-type and mutants flies could reveal the status of DC and MSCI in the germline (Figure 4). X chromosome down-regulation due to MSCI is expected to occur only in meiosis, a phase that is only completely present in wild-type testes. Hypertranscription of the X chromosome due to DC, in contrast, is expected to be found in both mutants and wild-type testis.
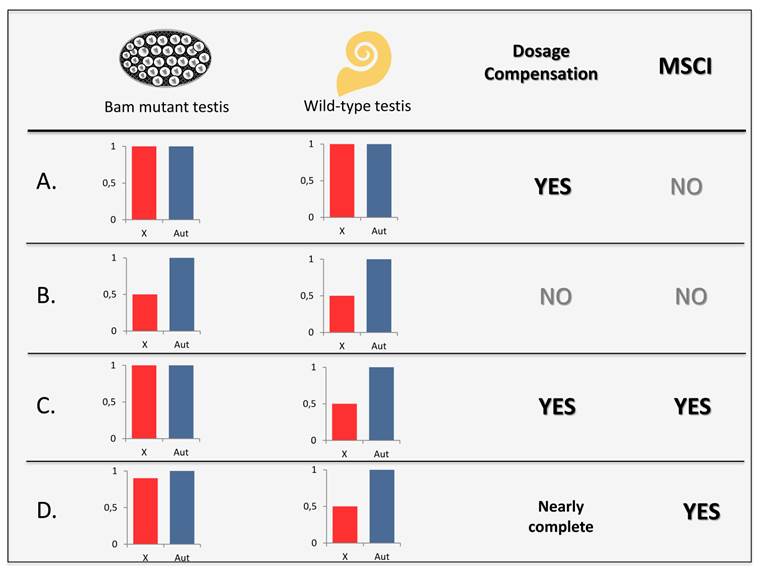
Microarray experiments found no global reduction in the expression of all X-linked genes in either wild-type or bcgn mutant testis [31]. This result suggested that the male germline is dosage compensated and MSCI is absent in meiotic cells as proposed in Figure 4A. However, the same group later analyzed RNAseq data [51] from another mutant, bam, to investigate the X: A median expression ratios [37]. While wild-type testes displayed significantly lower expression of X-linked genes (~65% of the autosomal expression), in testes from bam mutants, X-linked expression is much more similar and not significantly different from those observed for autosomes; (~90%) (Figure 4D) [37]. These results indicate not only that there is X chromosome DC in early mitotic cells but that MSCI exists, as X chromosome expression is even lower in wild-type testis [37]. Similar results were concomitantly observed by Meiklejohn et al. [33] when analyzing the same data [51]. This latter study found significant 10% reduction in the expression of bam mutant testis, suggesting some level of DC in mitotic cells [33]. The study also confirmed significantly larger expression reduction in the X chromosome in wild-type testis [33]. However, the authors concluded that the down regulation observed in differentiating spermatocytes was due to a previously unrecognized mechanism of X chromosome [33]. Very similar results were thus interpreted as different phenomena [33, 37]. The first group [37] interpreted the results as nearly complete DC and evidence for the existence of MSCI (Figure 4D), whereas the second group [33] suggested that incomplete DC and the existence of another previously undescribed mechanism of X chromosome down regulation better explained the observed results.
Recently, however, a reanalysis of microarray data [52] from an independent study of different bam mutant alleles found reduced (by ~50%) X-chromosome expression in both mutant and wild-type testes as proposed in Figure 4B (Table 2) by Meiklejohn and Presgraves [49]. This result suggests not only a lack of DC but also absence of MSCI. X chromosome expression reduction in wild-type testis would likely to be consequence of the lack of DC extended to later spermatogenic phases [49]. Microarray experiments produce expression signal for all genes regardless their real expression and therefore the application of probe filtering is a legitimate procedure to avoid effects from background hybridization. However, the exclusion of datasets with expression values lower than a certain threshold used in this study [49] might not be completely safe. Mutants in theory lack the expression of certain genes that are present in the wild-type flies. The removal of those genes from the experimental equation could minimize the differences between the two samples. The same study also selected probe sets showing the strongest signal intensity across all samples [49] which could bias the data by homogenizing the intensity values among wild-type and bam mutant testes.
In order to investigate if probe filtering could affect the X-autosome expression differences, I re-analyzed the microarray data from [52]. No probe selection or an alternative probe filtering provided results supporting a different scenario (Table 2). Bam mutants never present intense (50%) reduction of X-chromosome expression. More importantly, wild-type testes consistently present a larger X-chromosome expression reduction than mutants, in agreement with the hypothesis in which X inactivation in meiotic cells is an important player.
In summary, it is known that mutants should be used with caution to assess expression profile spermatogenesis as they present an aberrant spermatogenesis [49]. However, bam mutants proved to be a powerful experimental approach to expose the presence of DC and MSCI in spermatogenesis [33, 37] and this study. The majority of those studies showed significant lower expression of the X-linked genes in wild-type testis in comparison to mutants which supports the existence of MSCI. In addition, in mutant testes which are depleted of meiotic cells, X-linked and autosomal genes showed similar, or at least more similar, expression as expected for DC occurring in the germline.
Different analyses for expression data on bam mutant and wild-type testis [52]
| Microarray Data | |||
|---|---|---|---|
| Re-analysis in [49] | Re-analysis with all probes | Re-analysis with filterb | |
| Bam Mutant | |||
| Median A - Median Xc | 0.61 | -0.27 | 0.327 |
| P-valuea | NA | 0.84 | 0.036 |
| Wild-Type Testis | |||
| Median A - Median Xc | 0.45 | 0.24 | 0.614 |
| P-valuea | NA | 9.46e-8 | 2.65e-11 |
a. Mann-Whitney test
b. Filter to remove absent expression. Only probes with Affymetrix presence call in at least one replicate microarray experiment of either bam or wild-type testis were kept. Random selection of one probe for genes with multiple probes. NA stands for not available.
c. Medians were given in log2 values.
DC and MSCI investigation through mutant and wild-type testes. Expected levels of expression for X-linked and autosomal genes in bam or bgcn mutant and wild-type testes considering the presence or absence of DC and MSCI. A. In a male germline with DC and lack of MSCI, X-linked and autosomal genes would show the same level of expression in both mutant and wild-type testes. Scenario observed in [31]. B. For cases lacking DC and MSCI, expression of X-linked would be equivalent of half those from autosomal genes in all types of testis. C. In a germline with both DC and MSCI, only wild-type testes would display expression of X-linked genes as half as the autosomal genes expression level. D. Observed scenario in [33,37] and in this work, MSCI and nearly complete DC. Wild-type testis showed significantly greater reduction of X-linked genes expression than mutant testis.
Global Expression Analyses using Larval Testis
Spermatogenesis starts during the early development of Drosophila [40, 53]. First stage larval testes contain the first wave of germline cells undergoing differentiation, i.e. the products of successive divisions of single gonial cells that have not yet reached the final stages of sperm maturation [53] (Figure 5A and B). Second and third instar larvae are progressively enriched with spermatocytes whereas the production of post-meiotic cells and mature sperm mostly occurs during pupal stages and adults [53] (Figure 5B). Therefore, comparing the expression profile of X chromosome and autosomes in larval testes could be a valuable experimental approach to test the MSCI hypothesis in Drosophila. This approach was first claimed to provide evidence against MSCI [34].
Mikhaylova and Nurminsky [34] presented two major findings, which support their claim that there is no MSCI in Drosophila. First, testis-biased genes located both in the X chromosome and in the autosomes are transcriptionally activated during the meiotic stages of larval spermatogenesis [34]. If MSCI occurs in male meiosis, the transcriptional activation of X-linked genes should not be expected [34]. However, the method for selection of testis-biased genes is intrinsically a selection of male-meiosis-biased genes. Testis-biased genes are obtained from the comparison between testes and ovaries. As the bulk of testis expression comes from spermatocytes, 80% of testis-biased genes are actually meiotically expressed [29]. Therefore testis-biased genes, regardless their genomic location, are expected to be highly expressed in meiosis and probably activated in that phase regardless their chromosomal location.
Description of larval testis expression analysis used in [34]. A. Larvae with testis. B. Cell content in the testis of 2nd instar larvae. C. General experiment description and statistics. Each X-linked and autosomal gene here respectively represented by chrX1,...,chrXn and Aut1,..,Autn have their expression measured in 10 replicates. Variation among replicates represents 30% of the total variance. Nevertheless, only the mean of replicates were used to assess chromosomal expression differences described in (D). D. Box plot representing the distribution of expression intensity means of 10 replicates for all D. melanogaster transcript in a given development stage. X-linked and autosomal statistics are shown in orange and blue, respectively.
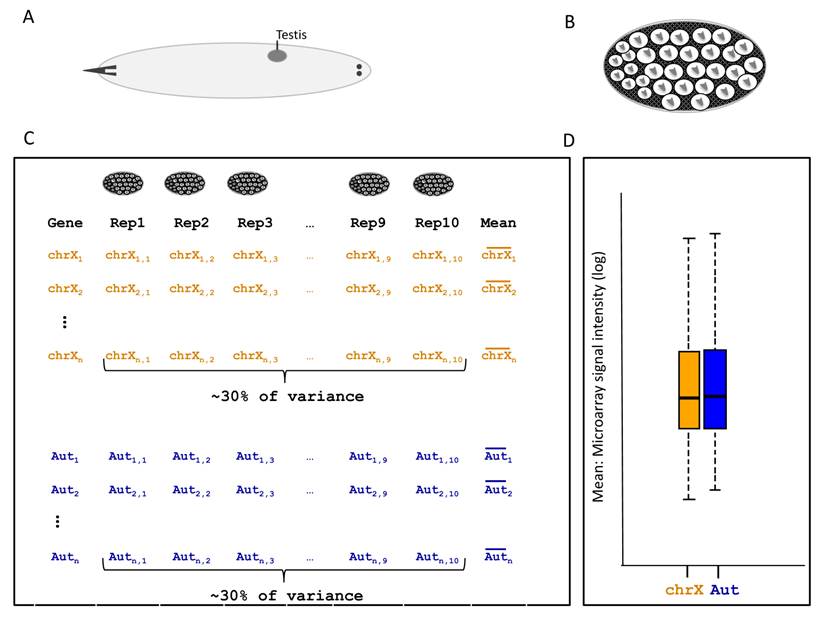
The second major finding against MSCI in this study comes from the global gene expression analysis in larval testis [34]. Genes on the X chromosome and autosomes were not differently expressed in testis in any of the larval stages analyzed [34] (Figure 5D). However, re-analyses of the data revealed a large experimental error between replicates that could compromise statistical power to detect expression differences [35]. More specifically, no significant difference in expression between X and autosomes was detected using comparisons of X chromosome and autosomal distributions for gene expression (Figure 5D). However, only the mean of the biological replicates (Figure 5C, column “Mean”) were used as data in Figure 5D. All the variance between biological replicate was disregarded in the first analysis [34]. The evaluation of variance (error) between replicates was much greater than in quality microarray experiments [29] (Figure 5C) which, if disregarded, may have affected the study's conclusions [35].
Analyses under proper statistical framework but still not accounting for replicate errors revealed significant down-regulation of the X chromosome with the increase of testis development and therefore enrichment with meiotic cells [35]. Although this finding was later acknowledged by all parties involved in the debate [36, 39], the discussion remained focused on the level of X-linked genes down-regulation and how far this pattern should be consider as a severe and global X chromosomal inactivation similar to the well-known MSCI in mammals [36,39,49].
It is not possible that an experimental error as described above [34-36] would allow us to evaluate the level of down-regulation of the X chromosome. This will only be clarified by redoing the biological experiments with smaller errors. By removing the noise caused by experimental errors, the down regulation of the X chromosome probably will be stronger. However, the currently observed X chromosome down regulation should not be neglected as an independent evidence for MSCI.
Final Remarks
Revisiting in detail all evidence regarding MSCI revealed that the negative argument in the debate is mostly based on erroneous interpretation of the same experimental data, often due to statistical misconceptions and errors and on the lack of knowledge about the methods used so far for MSCI detection in D. melanogaster. In order to obtain large amount of RNA previously required in global expression profiles, large-scale dissection created the sample mixed mitotic and meiotic samples [29]. In addition, it is unclear if the Y undergoes any down-regulation since the expressions of Y-linked genes during spermatogenesis have not been carefully analyzed in flies. Once those limitations were properly handled in statistical analysis, evidence in favor of MSCI becomes clear, but magnitude, range and the exactly timing in spermatogenesis are still interesting problems to investigate. A new era for high throughput of expressed sequence analyses and for single cell dissection has begun that potentially will allow the determination of the precise time and global range of MSCI in D. melanogaster.
Although there is no power to state the magnitude of MSCI in Drosophila, a lot of comparisons have been made to mammals MSCI that has been claimed to be stronger [33, 36, 39]. The comparison between the magnitude of the X-linked genes down regulation in Drosophila and in mammals is not meaningful for understanding MSCI evolutionary role. The real question should be that whether or not there is a significant signal of MSCI. If there is, then MSCI can play a role in evolution as even weak selection can have a significant effect in long evolutionary processes.
In fact, in this case, because of the differences of population sizes between organisms, a possible smaller level of MSCI in Drosophila and therefore an assumed weaker selection could have a similar role in evolution as the strong MSCI in mammals as the latest have much smaller population size. Indeed, compensatory meiotic expression of genes duplicated from the X chromosome to autosomes has been found for both mammals and Drosophila revealing the evolutionary outcomes of selection against X-linked genes expressed in males during the MSCI [28, 29].
Acknowledgements
I deeply thank Manyuan Long, Timothy L. Karr and Hedibert F. Lopes for the work done together and the innumerous discussions about MSCI. I greatly thank Nicholas Vankuren for scientific comments and proofreading of the manuscript. I thank Domitille Chalopin for taking the picture of the dissected apical tip shown in Figure 2A. I also thank John Parsch and Claus Kemkemer for discussion. MDV were supported by a National Institutes of Health grant (NIH R0IGM078070-01A1), the NIH ARRA supplement grant (R01 GM078070-03S1) and by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for the travel grant (2013/09714-6).
Competing Interests
The author has declared that no competing interest exists.
References
1. Lifschytz E, Lindsley DL. The role of the X-chromosome inactivation during spermatogenesis. Proc Natl Acad Sci. 1972;69:182-6
2. Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134:1823-31
3. McKee BD, Handel MA. Sex chromosomes, recombination, and chromatin conformation. Chromosoma. 1993;102:71-80
4. Richler C, Soreq H, Wahrman J. X inactivation in mammalian testis is correlated with inactive X-specific transcription. Nature Genet. 1992;2:192-5
5. Kelly WG, Schaner CE, Dernburg AF. et al. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479-92
6. Cabrero J, Teruel M, Carmona FD. et al. Histone H3 lysine 9 acetylation pattern suggests that X and B chromosomes are silenced during entire male meiosis in a grasshopper. Cytogenet Genome Res. 2007;119:135-42
7. Solari AJ. The behaviour of the XY pair in mammals. Int Rev Cytol. 1974;38:273-317
8. Fernandez-Capetillo O, Mahadevaiah SK, Celeste A. et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell. 2003;4:497-508
9. Turner JM, Mahadevaiah SK, Elliott DJ. et al. Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. J Cell Sci. 2002;115:4097-105
10. Namekawa S. H, Park P. J, Zhang L-F, et al. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660-7
11. Lee JT. Sex chromosome inactivation: The importance of pairing. Curr Biol. 2005;15:R249-52
12. Bean CJ, Schaner CE, Kelly WG. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat Genet. 2004;36:100-5
13. Turner JMA, Mahadevaiah SK, Fernandez-Capetillo O. et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41-7
14. McKee BD. Homolog pairing and segregation in Drosophila meiosis. Genome Dyn. 2009;5:56-68
15. Goñi B, Matsuda M, Yamamoto MT, Vilela CR, Tobari YN. Crossing over does occur in males of Drosophila ananassae from natural populations. Genome. 2012Jul;55(7):505-11
16. Vicoso B, Bachtrog D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature. 2013;499:332-5
17. Tao Y, Araripe L, Kingan SB. et al. A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol. 2007;5:e293
18. Wu C-I, Xu EY. Sexual antagonism and X inactivation - the SAXI hypothesis. Trends Genet. 2003;19:243-7
19. WR Rice. Sex chromosome and the evolution of sexual dimorphism. Evolution. 1984;38:735-42
20. Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537-40
21. Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854-9
22. Vibranovski MD, Zhang Y, Long M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897-903
23. Meisel RP, Han MV, Hahn MW. A Complex Suite of Forces Drives Gene Traffic from Drosophila X Chromosomes. Genome Biol Evol. 2009;0:176-88
24. Bai Y, Casola C, Feschotte C. et al. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007;8:R11
25. Parisi M, Nuttall R, Naiman D. et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697-700
26. Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14:2911-8
27. Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311-23
28. Potrzebowski L, Vinckenbosch N, Marques AC. et al. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80
29. Vibranovski MD, Lopes HF, Karr TL. et al. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731
30. Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:e273
31. Sturgill D, Zhang Y, Parisi M. et al. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238-41
32. Guioli S, Lovell-Badge R, Turner JM. Error-prone ZW pairing and no evidence for meiotic sex chromosome inactivation in the chicken germ line. PLoS Genet. 2012;8:e1002560
33. Meiklejohn CD, Landeen EL, Cook JM. et al. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 2011;9:e1001126
34. Mikhaylova LM, Nurminsky DI. Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome. BMC Biol. 2011;9:29
35. Vibranovski MD, Zhang YE, Kemkemer C. et al. Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the drosophila X chromosome. BMC Biol. 2012;10:49
36. Mikhaylova LM, Nurminsky DI. No severe and global X chromosome inactivation in meiotic male germline of Drosophila. BMC Biol. 2012;10:50
37. Deng X, Hiatt JB, Nguyen DK. et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43:1179-85
38. Kemkemer C, Hense W, Parsch J. Fine-scale analysis of X chromosome inactivation in the male germ line of Drosophila melanogaster. Mol. Biol. Evol. 2011;28:1561-3
39. Kemkemer C, Catalán A, Parsch J. 'Escaping' the X chromosome leads to increased gene expression in the male germline of Drosophila melanogaster. Heredity. 2014;112:149-55
40. Cooper KW. Normal spermatogenesis in Drosophila. In: (ed.) Demerec M. Biology of Drosophila. New York: Hafner. 1951:1-61
41. Kremer H, Hennig W, Dijkhof R. Chromatin organization in the male germ line of Drosophila hydei. Chromosoma. 1986;94:147-61
42. Pennisi E. Opening the way to gene activity. Science. 1997;275:155-7
43. Rastelli L, Kuroda MI. An analysis of maleless and histone H4 acetylation in Drosophila melanogaster spermatogenesis. Mech Dev. 1998;71:107-17
44. Hoyle HD, Hutchens JA, Turner FR. et al. Regulation of beta-tubulin function and expression in Drosophila spermatogenesis. Dev. Genet. 1995;16:148-70
45. Parsch J, Meiklejohn CD, Hauschteck-Jungen E. et al. Molecular evolution of the ocnus and janus genes in the Drosophila melanogaster subgroup. Mol Biol Evol. 2001;18:801-11
46. Baker BS, Gorman M, MarÌn I. Dosage compensation in Drosophila. Annu Rev Genet. 1994;28:491-521
47. Clark ME, Veneti Z, Bourtzis K. et al. The distribution and proliferation of the intracellular bacteria Wolbachia during spermatogenesis in Drosophila. Mech Dev. 2002;111:3-15
48. Hamada FN, Park PJ, Gordadze PR. et al. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289-94
49. Meiklejohn CD, Presgraves DC. Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline. Genome Biol Evol. 2012;4:1007-16
50. Gonczy P, Matunis E, DiNardo S. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development. 1997;124:4361-71
51. Gan Q, Chepelev I, Wei G. et al. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Research. 2010;20:763-83
52. Chen X, Lu C, Prado JR. et al. Sequential changes at differentiation gene promoters as they become active in a stem cell lineage. Development. 2011;138:2441-50
53. Lindsey DL, Tokuyasu KT. Spermatogenesis. In: (ed.) Ashbumer M, Wright TRF. The Genetics and Biology of Drosophila, Volume 2. London: Academic Press. 1980:226-294
Author contact
![]() Corresponding author: Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, São Paulo, Brazil 05508.
Corresponding author: Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, São Paulo, Brazil 05508.
Published 2014-6-1
