ISSN: 1839-9940
J Genomics 2015; 3:85-94. doi:10.7150/jgen.12975 This volume Cite
Research Paper
Whole Genome Sequence of the Soybean Aphid Endosymbiont Buchnera aphidicola and Genetic Differentiation among Biotype-Specific Strains
1. Department of Biology, Brandon University, Brandon, MB R7A 6A9, Canada.
2. Department of Entomology, The Ohio State University, OARDC, Wooster, OH 44691, USA.
Published 2015-10-5
Abstract
Endosymbiosis with microorganisms is common in insects, with more than 10% of species requiring the metabolic capabilities of intracellular bacteria for their nutrient acquisition. Aphids harbor an obligate mutualism with the vertically transferred endosymbiont, Buchnera aphidicola, which produces key nutrients lacking in the aphid's phloem-based diet that are necessary for normal development and reproduction. It is thought that, in some groups of insects, bacterial symbionts may play key roles in biotype evolution against host-plant resistance. The genome of Buchnera has been sequenced in several aphid strains but little genomic data is currently available for the soybean aphid (Aphis glycines), one of the most important pests of soybean in North America. In this study, DNA sequencing was used to assemble and annotate the genome sequence of the Buchnera A. glycines strain and to reconstruct phylogenetic relationships among different strains. In addition, we identified several fixed Buchnera SNPs between Aphis glycines biotypes that were avirulent or virulent to a soybean aphid resistance gene (Rag1). The results of this study describe the genetic and evolutionary relationships of the Buchnera A. glycines strain, and begin to define the roles of an aphid symbiont in host-plant resistance.
Keywords: DNA sequencing, host-plant resistance, de novo assembly Aphis glycines
Introduction
Insects are the most abundant and diverse animal class on earth, and are associated with a remarkable variety of symbiotic microorganisms. Their codiversification with intracellular bacterial symbionts (also called endosymbionts) has been well documented, including nearly all groups of phloem sap-sucking insects (1-3). Sap sucking insects often rely on their obligate mutualistic bacteria to synthesize nutrients that are deficient in their purely phloem diet but necessary to sustain normal development and reproduction (4, 5). One of the best studied cases of such symbiosis is between aphids and their obligate bacterial endosymbiont, Buchnera aphidicola (Proteobacteria: Enterobacteriaceae) (6-8).
The mutualism between aphid and Buchnera dates back over 200 million years and neither the insect nor its endosymbiont can survive independently (9). These vertically transmitted bacteria live within specialized bacteriocytes, aggregated to form the organ-like bacteriome located near the aphid midgut (6, 8). Aphids are dependent on Buchnera to produce some of the essential amino acids, vitamins, and sterols that are necessary for normal development and reproduction (10-15). In turn, the aphid provides Buchnera with nutrients, including nonessential amino acids and carbohydrates that are abundant in their phloem-based diet or produced by the host. Genomic evidence suggests that several amino acid biosynthetic pathways are shared between aphid and Buchnera, providing the aphid the ability to regulate the endosymbiont's metabolism (16).
No aphid species poses a greater threat to soybean yields in North America than the soybean aphid, Aphis glycines Matsumura (Hemiptera: Aphididae). Native to southeastern and eastern Asia, the species was first detected in Wisconsin in the summer of 2000 and has quickly spread throughout much of the North Central United States and Eastern Canada (17, 18). Host-plant resistance (referring to the plant's ability to resist damaging insect invasions) is often an effective method for controlling soybean pests, and at least five soybean aphid resistance genes (Rag1, Rag2, Rag3, Rag4, and Rag5) have been reported (19-26). However, biotypes of the soybean aphid have been identified in Midwest growing regions; biotypes are based on the ability to overcome host-plant resistance provided by one or more of these Rag genes. Biotype 2 can override the Rag1 gene (27), while biotype 3 can deride Rag2 resistance (28). Biotype 4 is capable of overcoming Rag1 and Rag2 defenses when expressed both singly and in concert (29). The mechanisms underlying biotype resistance are not well understood; however, recent work indicates Rag1 resistance may be associated with changes to the plant nutritional quality (30) or secondary plant metabolites (31), and elicits a transcriptional response typical of xenobiotic challenge in A. glycines (32). While most ongoing studies continue to focus on the plant and/or insect, one possibility is that the virulent phenotypes may be associated with aberrations in the insect microbiome, particularly the obligate symbiont, Buchnera (33). Recent work on Russian wheat aphids (Diuraphis noxia) and greenbugs (Schizaphis graminum) have revealed correlations between the symbiont profiles and biotypes (34, 35) but their genetic underpinnings have not yet been explored.
Genome sequences are currently available for several Buchnera strains derived from aphids of different tribes and subfamilies (36-39). The genome is small (<1 Mb) in comparison to other bacteria (40), having undergone reductive evolution involving the irreversible loss of genes and regulatory capabilities (37, 41). Many of the genes lost at the beginning of the symbiotic association functioned in gene transfer and/or recombination; thus, Buchnera has evolved independently in different host aphids, with little to no genetic exchange among strains (42). Consequently, phylogenetic relationships between aphids and Buchnera closely mirror one another across deep evolutionary divergences (43-46).
Transcript data has been accumulating for the Buchnera A. glycines strain (47, 48); however, the genome sequence and phylogenetic reconstructions are not yet available. Since Buchnera is not culturable, we performed next-generation DNA sequencing on whole body A. glycines to indirectly generate Buchnera sequence information. We assembled and functionally annotated the Buchnera genome, and reconstructed phylogenetic relationships. Under the premise that genetic differences in Buchnera may contribute to host-plant resistance, we carried out whole-genome resequencing of the Buchnera A. glycines strain in biotypes 1 and 2 to identify single nucleotide polymorphisms (SNPs) between them. We speculated that if SNPs were detected, they may be associated with genes involved in the aphid's nutrient synthesis and/or the counter-response to plant antiherbivory defense mechanisms (e.g. detoxification genes) - processes known to be mediated by microbial symbionts in the insect host. The results of this study characterize evolutionary and genetic associations of the Buchnera A. glycines strain and begin to define the roles of the aphid microbiome in host-plant resistance.
Materials and methods
Isolation of Buchnera DNA
Buchnera are obligatory symbiotic bacteria that have never successfully been cultured. Thus, we took advantage of next-generation sequencing technologies capability of generating sequence data for not only the aphid but the various associated microorganisms, including Buchnera. The A. glycines biotype 1 colony was established in 2008 at Ohio Agricultural Research and Development Center (OARDC) in Wooster, OH, using aphids from the University of Illinois' Soybean Aphid Biotype Stock Center. The biotype 2 colony was founded in 2005 from field collections in Ohio. Both colonies were periodically re-infested with aphids collected from natural populations. Subsequent to aphid collections, host plant resistance conferred by each biotype was validated using the Rag1 expressing soybean line 'A08-123074', and aphid susceptible soybean line 'SD01-76'. Biotype 2 was capable of surviving on the Rag1 plants (i.e. virulent), whereas biotype 1 was not (e.g. avirulent).
Whole body apterous aphids were simultaneously collected from each biotype and genomic DNA was extracted from the pools of 50 aphids using the E.Z.N.A® Tissue DNA Kit (Omega Bio-Tek, Norcross, GA). DNA (1 µg per sample) was used to generate an adaptor-ligated double-stranded DNA library for DNA sequencing using the TruSeq DNA PCR-Free Sample Prep Kit (Illumina, San Diego, CA) following the manufacturer's protocol. Quantification of DNA was done using the Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and quality was assessed using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). The samples (biotypes 1 and 2) were diluted to 18 nM and pooled to generate the multiplexed DNA library.
Illumina library synthesis and sequencing
The DNA library (36 fmoles) was sequenced on three flow cell lanes using the Illumina MiSeqTM2000 platform at the Ohio State University Molecular and Cellular Imaging Center. The mean library insert sequence length was 475 bp and both ends of the library were sequenced to generate 250 nt raw paired-end reads. Illumina Analysis Package CASAVA 1.8.2 was used to perform bcl conversion and demultiplexing. Image deconvolution and quality value calculations were performed using the Illumina GA pipeline v1.6.
Genome assembly and gene prediction
The 88.29 million raw paired-end reads of ≥240 bp were imported into CLC Genomics Workbench (v6.5.1, CLC Bio) and trimmed for quality and adapter indexes using the default settings (Ambiguous limit = 2, quality limit = 0.05). The remaining processed reads (87.53 million) were de novo assembled into scaffolded contigs of at least 1,000 bp using an algorithm based on de Bruijn graphs and the optimized parameters defined in Table S1. The final set of 40,643 contigs (mean length of 7,188 bp and N50 of 5,782 bp) was obtained by collapsing reads with ≥90% sequence similarity into clusters and retrieving the longest contig using CD-HIT (49). By mapping contigs against the nr database using BLAST2GO software (50), three contigs were identified as Buchnera aphidicola in origin (Buchnera Ag strain, BAg). The contigs corresponded to the circular chromosome and two circular plasmids. The raw sequence reads can be retrieved from the NCBI short sequence read archive under the accession number SRP045572 (Table 1).
To assess genome completeness, the preprocessed reads were mapped to each BAg contig using the resequencing function in CLC and the parameters described in Table S1. Since the BAg contigs are all circular, we were able to fill in any missing nucleotide sequence at each end by merging the overlapping sequence. Sequence alignment was also corroborated using this approach. Coverage analysis of the genome was conducted using the create statistics for target regions resequencing functions.
The Rapid Annotations based on Subsystem Technology (RAST) Genome Annotation Server v2.0 was used to annotate the identified Buchnera contigs. Functional annotation was done using the classic RAST scheme and GLIMMER-3 (Release70) gene caller. For comparison purposes, RAST was also used to annotate the Buchnera genome of the greenbug, Schizaphis graminum strain (BSg) (37).
Additional sequence of bacterial origin was identified in our de novo assembly. This included four contigs of the Wolbachia endosymbiont, as well as contigs of Escherichia, Halomonas, Lactobacillus, Rhodospirillum, and Streptomyces species. However, these contig sequences represented only a small fraction of their respective genomes.
NCBI accession numbers for the Buchnera short read data and genome sequence
| NCBI Resource | Accession Number |
|---|---|
| Small Read Archive (SRA) | SRP045572 |
| BioProject | PRJNA256101 |
| BioSample | SAMN02937548 (biotype 1) SAMN02937549 (biotype 1) SAMN02937550 (biotype 2) SAMN02937550 (biotype 2) |
| Genbank-Genomes | CP009253 (chromosome) CP009254 (pLeu) CP009255 (pTrp) |
Phylogenetic analysis
Single gene matrices and the concatenated sequence of 60 protein-coding genes shared by the Buchnera A. glycines strain and thirteen Buchnera strains from seven aphid species and deposited in the NCBI Bacterial Genomes FTP (accessed February 18, 2014) (Table 2) were used to reconstruct the phylogenetic associations among them. Maximum-Likelihood (ML) trees were constructed from both the individual and concatenated alignments using the CLC Phylogeny Module. The starting tree algorithm implemented the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) and rate variation (4 substitution rate categories, Gamma distribution parameter = 1.0). The 'Model Testing' tool in CLC was used for selection of the best-fit model of nucleotide substitution to be used for maximum likelihood phylogeny tree reconstruction. The tool employs four different statistical analyses to test the substitution models. The reliability of tree topologies was evaluated using 1000 bootstrap replicates. For each phylogeny, the relative substitution rates between pairs of Buchnera strains was calculated, using E. coli as an outgroup. Tree topologies were validated using only the first and second codon positions for tree inference, indicating mutations in the third codon positions were not saturated.
The thirteen Buchnera strains from seven aphid species and four tribes deposited in the NCBI Bacterial Genomes FTP (accessed February 18, 2014)
| Host Species | Common name | Subfamily | Tribe | Buchnera Straina | # Sequencesb |
|---|---|---|---|---|---|
| Acyrthosiphon kondoi | blue alfalfa aphid | Aphidinae | Macrosiphini | BAk | 559 |
| Acyrthosiphon pisum | pea aphid | Aphidinae | Macrosphini | BAp (TLW03) | 573 |
| BAp (JF98) | 477 | ||||
| BAp (JF99) | 590 | ||||
| BAp (5A) | 555 | ||||
| BAp (LL01) | 577 | ||||
| BAp (Tuc7) | 553 | ||||
| BAp (APS) | 564 | ||||
| Baizongia pistaciae | N/A | Eriosomatinae | Fordini | BBp | 504 |
| Cinara cedri | cedar aphid | Lachninae | Eulachinini | BCc | 357 |
| Cinara tujafilina | cypress pine aphid | Lachninae | Eulachinini | BCt | 360 |
| Schizaphis graminum | greenbug | Aphidinae | Aphidini | BSg | 546 |
| Uroleucon ambrosiae | brown ambrosia aphid | Aphidinae | Macrosiphini | BUa | 529 |
aThe Acyrthosiphon pisum strain is denoted in parentheses.
bThe number of sequences were derived from the .ffn files in the NCBI FTP database.
Sequence polymorphism analysis
Single nucleotide polymorphisms (SNPs), insertions, and deletions in the Buchnera genome (see above) were identified between Aphis glycines biotypes 1 and 2 using the quality variant detection re-sequencing function (based on the Neighborhood Quality Standard (NQS) algorithm) in CLC Bio. Only polymorphisms between biotypes with read coverage of ≥25 valid reads were considered. Mutations in coding regions that putatively result in non-synonymous changes were identified using the GLIMMER-3 predicted protein-coding sequences.
Results
Genome assembly and annotation
In this study we assembled the complete genome of Buchnera Ag strain (BAg). To do this, we carried out DNA sequencing of its host aphid species (Aphis glycines). De novo assembly of preprocessed reads and subsequent resequencing analyses produced a complete BAg genome of 638,852 bp, with 25.6% GC content. It consisted of a 628,164 bp chromosome, pLeu plasmid (7,639 bp), and pTrp plasmid (3,049 bp). The chromosome and plasmid sequences have been deposited in the NCBI GenBank archive under the accession numbers CP009253-CP009255 (Table 1).
Average coverage of the chromosome (x = 846.9) and pLeu (x = 777.3) were similar, whereas pTrp had roughly twelve-fold higher coverage (x = 10,244.1). The BAg chromosome and plasmid sizes and sequence alignments were coanalyzed and corroborated using de novo assembly algorithms of CLC Bio and Velvet (51) in the MCIC-Galaxy automated pipeline (52).
The RAST Prokaryotic Genome Annotation Server was used to annotate the genes, which implements a 'highest confidence first' propagation strategy (see methods) and the GLIMMER-3 gene prediction scheme. A total of 593 putative protein-coding genes and 35 RNAs were identified on the chromosome, as well as 7 and 3 protein-coding genes on the pLeu and pTrp plasmids, respectively (no RNAs) (Table S2). Thirteen of the chromosome genes had predicted amino acid sequences of <50 aa and could not be assigned function. Functional categorization of the protein-coding genes is displayed in Fig. 1. Approximately 83% (n = 503) of genes could be assigned to one or more functional groups, based on current Buchnera subsystem annotation.
Table 3 shows the comparison of the BAg genome with selected genomes of Buchnera strains of other aphid species, which was adapted from the KEGG genome database (53) and BuchneraBASE (54). The genome is comprised of 88.5% coding sequence and 25.6% GC content. The BAg genome contained the largest number of protein coding genes, with 603 predicted for the chromosome and plasmids combined. It also contained the smallest number of RNAs, even in comparison to the significantly reduced BBp and BCc genomes. This was primarily due to an absence of sRNAs in BAg, whereas all of the other strains had a minimum of two sRNAs. While it remains difficult to identify sRNAs by sequence inspection, pairwise analyses of the BAg genome with sRNA sequences derived from the other strains was unable to uncover any putative sRNAs. It is unclear whether there are any functional implications for the bacteria or aphid host related to the absence of sRNAs but it could be related to the extreme specialization of A. glycines on two seasonal hosts.
Comparisons of Buchnera aphidicola genomes derived from A. glycines (BAg) and selected other aphid species. The data presented was adapted from the KEGG genome database (Kanehisa and Goto 1999) and BuchneraBASE (Prickett et al. 2006)
| Feature | BAg | BBp | BAp (APS) | BSg | BCc | |
|---|---|---|---|---|---|---|
| Genome | ||||||
| Genome Size (bp) | 638,852 | 618,379 | 655,725 | 653,001 | 422,434 | |
| GC content (%) Protein-coding genes | 25.6 603 | 25.3 507 | 26.4 574 | 26.3 555 | 20.2 362 | |
| Chromosome | ||||||
| Size (bp) | 628,164 | 615,980 | 640,681 | 641,454 | 416,380 | |
| Protein-coding genes1 | N/A (593) | 504 (510) | 564 (578) | 546 (600) 37 | 357 (371) | |
| RNAs | 35 | 37 | 36 | 37 | ||
| Plasmids | ||||||
| Number | 2 | 1 | 2 | 2 | 1 | |
| Total size (bp) | 10,688 | 2,399 | 15,044 | 11,547 | 6,054 | |
| Protein-coding genes | 10 | 3 | 10 | 9 | 5 |
1The number in parenthesis indicates the predicted number of protein-coding genes using the Rapid Annotations based on Subsystem Technology (RAST) Genome Annotation Server v2.0.
Functional categorization of the protein-coding genes in the Buchnera aphidicola genome of A. glycines
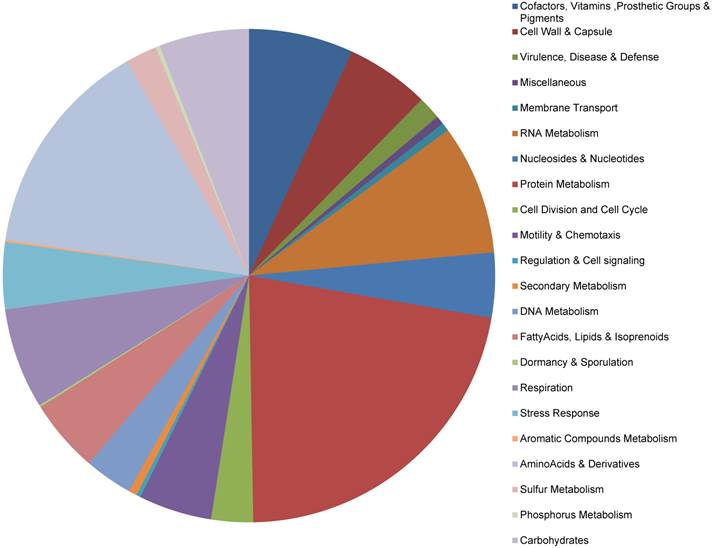
Functional comparison between BAg and BSg
Since the BAg and BSg belong to the same tribe and are the most closely related Buchnera strains currently available in the NCBI genome database, we implemented pairwise functional analyses between strains using the RAST annotation for both BAg and BSg. Roughly 85% of the BAg and BSg chromosome sequence overlapped and these regions were ~84% similar. The chromosome sequence of all other Buchnera strains had less than 70% overlap and 82% similarity. The BAg plasmid sequences had slightly less overlap (78%) and similarity (81%) with BSg plasmids. The gene order was identical but gene composition differed. A subset of genes was present in either BAg or BSg but absent in the other strain (present only in BAg: bioH, cmk, cysQ, hemD, lig, mltE, yfaE, ydgO, ydiC, and yhiQ; present only in BSg: YidD, YjeK, SohB, topA, mutS, mltA, rpmJ, ygfA, and MutL). There were also a two instances where a single-copy BSg gene (mrsA, and murE) was found in two adjacent copies in BAg; however, one gene copy was dramatically reduced in size and likely reflects remnant sequence of the functional gene.
Phylogenetic reconstruction of BAg
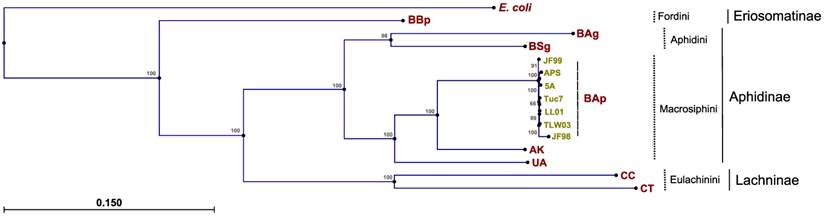
Phylogenetic relationships between Buchnera strains were explored previously (45, 46), but these studies did not incorporate sequence data from BAg. As expected, the topologies derived from the maximum likelihood analysis of 60 concatenated protein-coding Buchnera genes placed the strains into distinct clades corresponding to current aphid tribe classifications (maximum log likelihood of -485470.99; transition/transversion ratio of 1.58) (Fig. 2). Phylogenies from the single-gene datasets were consistent with the concatenated analysis, with the Fordini, Eulachinini, and Macrosphini tribes placed in monophyletic clades. For most genes, the Aphidini tribe formed a distinct clade. There were six instances (rpmI, rpmF, csrA, metE, rplT, and groeS) where BSg or BAg clustered more closely with the Macrosphini clade than to its fellow Aphidini member, which points towards potential issues in reconstructing symbiont phylogenies based on only one or a few genes.
BAg sequence differentiation between aphid biotypes
Genome size and gene composition was identical for the BAg genomes of A. glycines biotypes 1 and 2. The quality variant detection re-sequencing function in CLC Bio was used to search for nucleotide polymorphisms, insertions, and deletions between the biotypes in the BAg genome. A total of 19 fixed homozygous polymorphisms were identified (Table 4). All of the polymorphisms were SNPs and most were detected on the chromosome (n = 17). The SNPs were spread throughout the chromosome, with 16 of the SNPs located in 15 protein-coding genes. Of interest, nine of the SNPs resulted in putative non-synonymous mutations, which are more likely to affect protein structure and/or function than synonymous changes (55, 56). Moreover, four of the SNPs putatively changed the chemical composition (i.e. polarity and/or pH) of the encoded amino acids. Several heterozygous sites were also identified on the chromosome (n = 14) and plasmids (n = 4), which may be characteristic of the within biotype diversity.
Phylogenetic reconstructions of Buchnera aphidicola A. glycines strain, derived from the maximum likelihood analysis of 60 concatenated protein-coding genes
The 19 fixed homozygous nucleotide polymorphisms identified in the Buchnera aphidicola A. glycines genome between A. glycines biotypes 1 and 2
| Genomic Location | Position (bp) | Biotype 1a | Biotype 2b | Gene | Mutation | AA (B1/B2)c | |
|---|---|---|---|---|---|---|---|
| Chromosome | 31,438 | A | C | metE | NS | Q/H | |
| Chromosome | 45,040 | G | A | rplA | S | G | |
| Chromosome | 79,764 | G | T | fliI | S | G | |
| Chromosome | 131,494 | G | A | pheS | NS | V/I | |
| Chromosome | 176,777 | T | G | N/A | N/A | N/A | |
| Chromosome | 254,880 | C | A | N/A | N/A | N/A | |
| Chromosome | 311,966 | A | C | bioD | NS | S/Ac | |
| Chromosome | 362,217 | A | G | pyrC | NS | T/Ac | |
| Chromosome | 381,331 | A | G | ycfH | S | A | |
| Chromosome | 448,664 | A | G | ispD | NS | K/Ec | |
| Chromosome | 455,674 | G | A | cysI | NS | T/S | |
| Chromosome | 496, 810 | G | A | thiL | S | L | |
| Chromosome | 502,106 | G | T | yajR | S | G | |
| Chromosome | 538,357 | A | G | RpsD | S | D | |
| Chromosome | 584,826 | C | A | N/A | N/A | N/A | |
| Chromosome | 598,886 | T | C | amiB | NS | I/Tc | |
| Chromosome | 612,816 | A | C | hemD | NS | I/L | |
| pLeu | 4,731 | C | T | leuA | S | E | |
| pLeu | 7,289 | C | A | leuA | NS | E/D | |
| pTrp | ---------- | ---------- | ---------- |
aAvirulent biotype.
bVirulent biotype for RAG1 resistance.
cAmino acid changes to different polarities and/or pH levels.
Discussion
Symbiosis with microorganisms occurs often in insects, with members of nearly all groups of sap-sucking insects requiring the metabolic capabilities of intracellular bacteria for normal development and reproduction. This includes the soybean aphid (A. glycines), which is one of the most important pests in soybean growing regions of the Midwest. All aphid species harbor an evolutionarily ancient obligate symbiosis with Buchnera aphidicola, which produces some of the nutrients that are deficient in the aphid's purely phloem diet. The genomes of several Buchnera strains have been sequenced (36-39), and their resulting small size strongly suggests that the bacteria have undergone degenerative evolution from its free living ancestral form. We were able to isolate and assemble de novo the chromosome and plasmid sequences of the Buchnera A. glycines strain (BAg) from whole body aphids, highlighting the effectiveness of next-generation sequencing in sequencing host-associated microorganisms.
The genome size of BAg was ~639 Kb, which is smaller than the fellow Aphidinae A. kondoi, A. pisum, and S. graminum, but considerably larger than members of the Lachninae subfamily (e.g. C. cedri and C. tujafilina). Most of the 603 predicted BAg genes could be assigned a function, and over half are involved in amino acid and protein metabolism, indicative of Buchnera's integral role in the nutrient biosynthetic pathways. The BAg genome contained the most protein coding genes of any Buchnera genome currently available. However, the number of predicted genes is likely a bit overestimated since 1) our annotation scheme did not detect pseudogenes; and 2) the predicted amino acid sequences of several hypothetical genes, absent from other Buchnera strains, were short (<50 aa) and probably not functional. Genome coverage of pTrp was substantially elevated, suggesting a greater copy number of the plasmid relative to the chromosome and pLeu in Buchnera cells. In Buchnera of other Aphidini members, anthranilate synthase is the limiting enzyme of the tryptophan biosynthetic pathway, and its coding genes (trpEG) are localized on pTrp (57-59). The remaining genes in the pathway are located on the chromosome (9). Buchnera is consequently capable of controlling the ratio of trpEG relative to chromosomal genes by modulating the pTrp copy number (57, 60). The BAg plasmid contained two copies of trpEG, which comprises the majority of the sequence. In addition, genes encoding the entire leucine biosynthetic pathway (leuABCD) are localized on pLeu (61), and BAg has one copy of each gene. The copy number of these plasmids differs among aphid species (57, 62-64), presumably due to adaptive modifications related to the varying nutritional requirements of the insect hosts. However, the disparity in the ratios of pTrp and pLeu to chromosome copy number is considerably greater for BAg than for the other Buchnera strains, and may suggest that A. glycines shows greater specificity for endosymbiont-derived essential amino acids.
The BAg genome was substantially diverged from the other sequenced strains, with <85% nucleotide similarity to Buchnera strains of its fellow Aphidini tribe member and the most phylogenetically related strain currently available in the NCBI Bacterial Genomes FTP, Schizaphis graminum (BSg). Gene order was identical between strains but there were differences in chromosomal gene composition. Most notable were genes encoding proteins involved in DNA replication and repair that were present only in BAg (topA, mutS, and mutL) or BSg (lig), indicating significant differences exist at the cellular level. The BAg genome also contained several genes functioning in building block biosynthesis (bioH, cmk, cysQ, and hemD), sulfur metabolism (ydiC and yfaE), and oxidative-reduction (ydgO), which were missing in BSg, suggesting BAg may retain some metabolic pathways that have since lost functionality in BSg. Conversely, BSg retained genes involved in macromolecule degradation (SohB) and posttranscriptional regulation (YjeK) that were no longer found in BAg. Also of interest, the BAg and BSg genomes contained different isoforms of the murein transglycosylase encoding gene (mltA and mltE, respectively). The considerable differences in nucleotide sequence and gene composition between Buchnera strains was expected since they have likely had little to no genetic exchange since the time of divergence of their common Aphidini ancestor (42).
Associations between biotype and host-plant resistance are common for species within the family Aphidiae (65, 66). A key attribute of this group, which may enhance its biotype adaptability relative to other insect pest species, is their obligate mutualism with Buchnera. Endosymbionts can be important mediators of direct and indirect interactions between herbivorous insects and their host plants, and could potentially contribute to biotype evolution or host-race formation of some insects due to their intricate role in nutrient synthesis (67, 68). A 2010 study found constitutive differences in amino acid composition between Rag1 and non-Rag1 soybean leaves, suggesting the resistance gene impacts nutritional quality of the plant in some capacity (30). In turn this may select for Buchnera A. glycines strain genotypes capable of overcoming the deficiencies in amino acid content in Rag1 plants. Proteome analysis revealed considerable differences in Buchnera expression patterns between potato aphid clones (Macrosiphum euphorbiae) with differing levels of virulence reared on resistant and susceptible tomato plants, suggesting the endosymbiont may influence the insect adaptation to host plant resistance (69). Most of the putative BAg nonsynonymous mutations identified between A. glycines biotypes 1 (avirulent) and 2 (virulent) bring about amino acid changes in proteins involved in amino acid activation (pheS) and the biosynthesis of amino acids (metE and leuA), cofactors (bioD, hemD, and ispD), and macromolecules (amiB), all of which could conceivably alter the nutrient profiles afforded by the different biotypes. Moreover, the genes bioD and ispD contain SNPs which change the chemical composition (i.e. polarity and pH) of the encoded amino acids, and could produce major changes to protein structure and stability (70).
Endosymbionts represent an important but often overlooked trophic level in aphid-plant interactions, and may contribute to the insect's differential adaptation to host plant resistance. Several A. glycines resistance genes have been reported and multiple aphid biotypes have been identified based on the ability to overcome host-plant resistance provided by these Rag genes. In this study, we carried out next-generation DNA sequencing to isolate, assemble, and annotate the genome of BAg from whole body aphids, as well as reconstruct the phylogenetic associations among different Buchnera strains. As expected, the BAg genome is small due to reductive evolution, comprised of ~639 Kb on one circular chromosome and two plasmids. While the genome is genetically distinct and differs in gene composition from other Buchnera strains, it forms a distinct clade with the other member of the Aphidini tribe. We detected several fixed, nonsynonymous BAg mutations between avirulent and virulent biotypes for Rag1 resistance, most of which could potentially impact the nutrient profiles afforded to the different biotypes. Future population genomics studies are needed to determine whether the identified SNPs represent fixed differences between biotypes that are consistent across multiple geographical locations. Additional targeted genetic, metabolomics, and proteomic studies will be needed to determine whether the genetic changes influence nutrient synthesis by Buchnera and contribute to aphid biotype evolution to overcome Rag1 resistance. Regardless, our results demonstrate that sequence divergence among Buchnera populations can occur quite rapidly, and could dramatically alter aspects of the aphid adaptability.
Supplementary Material
Table S1.
Acknowledgements
Soybean aphids used in this study were initially supplied from the Soybean Aphid Biotype Stock Center by Dr. Curt Hill at the University of Illinois.
Funding
This research was supported by The Ohio State University, Center for Applied Plant Sciences, Ohio Soybean Council, North Central Soybean Research Program, and USDA-North Central IPM Center.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Allen JM, Reed DL, Perotti MA, Braig HR. Evolutionary relationships of "Candidatus Riesia spp, " endosymbiotic enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol. 2007;73:1659-1664
2. Conord C, Despres L, Vallier A, Balmand S, Miquel C. et al. Long-term evolutionary stability of bacterial endosymbiosis in curculionoidea: additional evidence of symbiont replacement in the dryophthoridae family. Mol Biol Evol. 2008;25:859-868
3. Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165-190
4. Moran NA, Telang A. Bacteriocyte-associated symbionts of insects - a variety of insect groups harbor ancient prokaryotic endosymbionts. Bioscience. 1998;48:295-304
5. Frago E, Dicke M, Godfray HCJ. Insect symbionts as hidden players in insect-plant interactions. Trends Ecol Evol. 2012;27:705-711
6. Buchner P. Endosymbiosis of animals with plant microorganisms. New York, USA: John Wiley and Sons Ltd. 1965
7. Munson MA, Baumann P, Kinsey MG. Buchnera gen.nov and Buchnera-Aphidicola sp.nov, a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol. 1991;41:566-8
8. Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155-189
9. Baumann P, Baumann L, Lai CY, Rouhbakhsh D, Moran NA. et al. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55-94
10. Nakabachi A, Ishikawa H. Differential display of mRNAs related to amino acid metabolism in the endosymbiotic system of aphids. Insect Biochem Molec. 1997;27:1057-1062
11. Moran NA, Dunbar HE, Wilcox JL. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 2005;187:4229-4237
12. Moran NA, Degnan PH. Functional genomics of Buchnera and the ecology of aphid hosts. Mol Ecol. 2006;15:1251-1261
13. Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218-229
14. Gunduz EA, Douglas AE. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. P Roy Soc B-Biol Sci. 2009;276:987-991
15. Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. P Natl A Sci USA. 2011;108:2849-2854
16. Wilson ACC, Ashton PD, Calevro F, Charles H, Colella S. et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 2010;19:249-258
17. Wu ZS, Schenk-Hamlin D, Zhan WY, Ragsdale DW, Heimpel GE. The soybean aphid in China: a historical review. Ann Entomol Soc Am. 2004;97:209-218
18. Venette RC, Ragsdale DW. Assessing the invasion by soybean aphid (Homoptera: Aphididae): Where will it end? Ann Entomol Soc Am. 2004;97:219-226
19. Hill CB, Li Y, Hartman GL. A single dominant gene for resistance to the soybean aphid in the soybean cultivar Dowling. Crop Sci. 2006;46:1601-1605
20. Hill CB, Li Y, Hartman GL. Soybean aphid resistance in soybean Jackson is controlled by a single dominant gene. Crop Sci. 2006;46:1606-8
21. Li Y, Hill CB, Carlson SR, Diers BW, Hartman GL. Soybean aphid resistance genes in the soybean cultivars Dowling and Jackson map to linkage group M. Mol Breeding. 2007;19:25-34
22. Mian MAR, Hammond RB, Martin SKS. New plant introductions with resistance to the soybean aphid. Crop Sci. 2008;48:1055-1061
23. Zhang GR, Gu CH, Wang DC. Molecular mapping of soybean aphid resistance genes in PI 567541B. Theor Appl Genet. 2009;118:473-482
24. Zhang GR, Gu CH, Wang DC. A novel locus for soybean aphid resistance. Theor Appl Genet. 2010;120:1183-1191
25. Kim K.S, Hill CB, Hartman GL, Hyten DL, Hudson ME. et al. Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theor Appl Genet. 2010;121:599-610
26. Jun TH, Mian MAR, Michel AP. Genetic mapping revealed two loci for soybean aphid resistance in PI 567301B. Theor Appl Genet. 2012;124:13-22
27. Kim K.S, Hill CB, Hartman GL, Mian MAR, Diers BW. Discovery of soybean aphid biotypes. Crop Sci. 2008;48:923-8
28. Hill CB, Crull L, Herman TK, Voegtlin DJ, Hartman GL. A New Soybean Aphid (Hemiptera: Aphididae) Biotype Identified. J Econ Entomol. 2010;103:509-515
29. Alt J, Ryan-Mahmutagic M. Soybean aphid biotype 4 identified. Crop Sci. 2013;53:1491-5
30. Chiozza MV, O'Neal ME, MacIntosh GC. Constitutive and induced differential accumulation of amino acid in leaves of susceptible and resistant soybean plants in response to the soybean aphid (Hemiptera: Aphididae). Environ Entomol. 2010;39:856-864
31. Li Y, Zou JJ, Li M, Bilgin DD, Vodkin LO. et al. Soybean defense responses to the soybean aphid. New Phytol. 2008;179:185-195
32. Bansal R, Mian MAR, Mittapalli O, Michel AP. RNA-Seq reveals a xenobiotic stress response in the soybean aphid, Aphis glycines, when fed aphid-resistant soybean. BMC Genomics. 2014;15:972
33. Wenger JA Michel AP. Implementing an evolutionary framework for understanding genetic relationships of phenotypically defined insect biotypes in the invasive soybean aphid (Aphis glycines). Evol App. 2013;6:1041-1053
34. Pinheiro P, Bereman MS, Burd J, Pals M, Armstrong S. et al. Evidence of the biochemical basis of host virulence in the greenbug aphid, Schizaphis graminum (Homoptera: Aphididae). J Proteome Res. 2014;13:2094-2108
35. Anathakrishnan R, Sinha DK, Murugan M. et al. Comparative gut transcriptome analysis reveals differences between virulent and avirulent Russian wheat aphids, Diuraphis noxia. Arthropod-Plant Inte. 2014;8:79-88
36. Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp APS. Nature. 2000;407:81-6
37. Tamas I, Klasson L, Canback B, Naslund AK, Eriksson AS. et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376-9
38. van Ham RC, Kamerbeek J, Palacios C, Rausell C, Abascal F. et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100:581-6
39. Perez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M. et al. A small microbial genome: The end of a long symbiotic relationship? Science. 2006;314:312-3
40. Thomas GH, Zucker J, Macdonald SJ, Sorokin A, Goryanin I. et al. A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Syst Biol. 2009:3
41. Moran NA, Plague GR, Sandstrom JP, JWilcox JL. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc Natl Acad Sci USA. 2003;100:14543-14548
42. The International Aphid Genome Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313
43. Moran NA, Munson MA, Baumann P, Ishikawa H. A Molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. P Roy Soc B-Biol Sci. 1993;253:167-171
44. Munson MA, Baumann P, Clark MA, Baumann L, Moran NA. et al. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of 4 aphid families. J Bacteriol. 1991;173:6321-6324
45. Perez-Brocal V, Gil R, Moya A, Latorre A. New insights on the evolutionary history of aphids and their primary endosymbiont Buchnera aphidicola. Int J Evol Biol. 2011;2011:250154
46. Novakova E, Hypsa V, Klein J, Foottit RG, von Dohlen CD. et al. Reconstructing the phylogeny of aphids (Hemiptera: Aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol Phylogenet Evol. 2013;68:42-54
47. Bai XD, Zhang W, Orantes L, Mittapalli O. et al. Combining next-generation sequencing strategies for rapid molecular resource development from an invasive aphid species, Aphis glycines. PLoS ONE. 2010;5:e11370
48. Liu SJ, Chougule NP, Vijayendran D, Bonning BC. Deep sequencing of the transcriptomes of soybean aphid and associated endosymbionts. PLoS ONE. 2012;7:e45161
49. Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658-9
50. Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420-3435
51. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821-9
52. Goecks J, Nekrutenko A, Taylor J, Galaxy T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86
53. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30
54. Prickett MD, Page M, Douglas AE, Thomas GH. BuchneraBASE: a post-genomic resource for Buchnera sp APS. Bioinformatics. 2006;22:641-642
55. Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198-200
56. Rudd MF, Williams RD, Webb EL, Schmidt S, Sellick GS. et al. The predicted impact of coding single nucleotide polymorphisms database. Cancer Epidemiol Biomarkers Prev. 2005;14:2598-2604
57. Lai CY, Baumann L, Baumann P. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci USA. 1994;91:3819-3823
58. Rouhbakhsh D, Lai CY, von Dohlen CD, Clark MA, Baumann L. et al. The tryptophan biosynthetic pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated anthranilate synthase (trpEG) within the aphididae. J Mol Evol. 1996;42:414-421
59. Lai CY, Baumann P, Moran NA. Genetics of the tryptophan biosynthetic pathway of the prokaryotic endosymbiont (Buchnera) of the aphid Schlechtendalia chinensis. Insect Mol Biol. 1995;4:47-59
60. Munson MA, Baumann P. Molecular cloning and nucleotide sequence of a putative trpDC(F)BA operon in Buchnera aphidicola (endosymbiont of the aphid Schizaphis graminum). J Bacteriol. 1993;175:6426-6432
61. Bracho AM, Martinez-Torres D, Moya A, Latorre A. Discovery and molecular characterization of a plasmid localized in Buchnera sp. bacterial endosymbiont of the aphid Rhopalosiphum padi. J Mol Evol. 1995;41:67-73
62. Lai CY, Baumann P, Moran NA. The endosymbiont (Buchnera sp.) of the aphid Diuraphis noxia contains plasmids consisting of trpEG and tandem repeats of trpEG pseudogenes. Appl Environ Microbiol. 1996;62:332-9
63. Thao ML, Baumann L, Baumann P, Moran NA. Endosymbionts (Buchnera) from the aphids Schizaphis graminum and Diuraphis noxia have different copy numbers of the plasmid containing the leucine biosynthetic genes. Curr Microbiol. 1998;36:238-240
64. Plague GR, Dale C, Moran NA. Low and homogeneous copy number of plasmid-borne symbiont genes affecting host nutrition in Buchnera aphidicola of the aphid Uroleucon ambrosiae. Mol Ecol. 2003;12:1095-1100
65. Smith C. Plant resistance to arthropods. Dordrecht, NLD: Springer Press. 2005
66. van Emden HF. Aphids as crop pests. Cambridge, UK. 2007
67. Simon JC, Carre S, Boutin M, Prunier-Leterme N, Sabater-Mun B. et al. Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc Biol Sci. 2003;270:1703-1712
68. Chiel E, Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Katzir N. et al. Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull Entomol Res. 2007;97:407-413
69. Francis F, Guillonneau F, Leprince P, De Pauw E, Haubruge E. et al. Tritrophic interactions among Macrosiphum euphorbiae aphids, their host plants and endosymbionts: Investigation by a proteomic approach. J Insect Physiol. 2010;56:575-585
70. Volkenstein MV. Coding of polar and non-polar amino-acids. Nature. 1965;207:294-5
Author contact
![]() Corresponding author: Bryan J. Cassone, Brandon University, Department of Biology, Brandon, MB, R7A 6A9. cassonebca; 204-727-7333 (Ph)
Corresponding author: Bryan J. Cassone, Brandon University, Department of Biology, Brandon, MB, R7A 6A9. cassonebca; 204-727-7333 (Ph)

 Global reach, higher impact
Global reach, higher impact