ISSN: 1839-9940Journal of Genomics
J Genomics 2020; 8:16-20. doi:10.7150/jgen.42095 This volume Cite
Research Paper
Isolation and genomic analysis of 11-aminoundecanoic acid-degrading bacterium Pseudomonas sp. JG-B from nylon 11 enrichment culture
Abstract
Nylon 11 is a polymer synthesized from 11-aminoundecanoic acid, and widely used in commercial manufacturing. In this study, we describe the isolation of the first organism capable of metabolizing 11-aminoundecanoic acid from nylon 11 enrichment culture. The strain shows rapid growth on 11-aminoundecanoic acid as a sole source of carbon, nitrogen, and energy. Furthermore, the genome sequence of strain JG-B was deciphered and shown to belong to genus Pseudomonas. Many genes encoding putative extracellular hydrolases, as well as homologues of nylon 6 hydrolases (NylB and NylA) were identified, suggesting the metabolic versatility and possibility that this organism could also depolymerase nylon 11 polymers.
Keywords: Nylon 11, biodegradation, genome, Pseudomonas, 11-aminoundecanoic acid, hydrolases.
Introduction
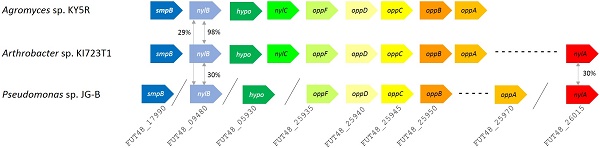
Nylon 11 is a common synthetic bioplastic that is used in the manufacture of a wide range of commercial products. Nylon 11 is synthesized by polymerization of 11-aminoundecanoic acid, which is derived from castor oil (Figure 1) [1]. Polymers form extensive inter-polymer interactions, which result in the crystalline structure and strength desired for many commercial materials. These structures, on the other hand, make polymers largely resistant to biotic and abiotic degradation. Therefore, when accidentally released into the environment, they remain intact for a long period of time and pose a significant threat to various organisms.
As a safe and cost-effective means to remediate plastics in the environment, biodegradation has attracted attention in recent years. Biodegradation of polyesters (e.g. polyethylene, polyurethane, and poly(ethylene terephthalate) (PET)) and polyamides (e.g. nylons) by bacteria and fungi species have been investigated in the last couple of decades [2-5]. Metabolic degradation of synthetic and biological polymers proceeds in two steps: extracellular fragmentations of polymers and then cellular metabolism of monomeric units.
Chemical structure of 11-aminoundecanoic acid and nylon 11.
Enzymes that hydrolyze synthetic polymers (plastics) have been identified [6, 7]. Hydrolases, including esterases, lipases and cutinase hydrolyze ester linkage of PET [8-10]. Extracellular poly(L-lactic acid) (PLA) depolymerase degrades PLA into lactic acid [7]. Studies with nylon 6 polymerization by-products identified three enzymes that are involved in microbial degradation of a variety of nylon 6 oligomers, namely 6-aminohexanoate-cyclic-dimer hydrolase (EI), 6-aminohexanoate-dimer hydrolase (EII), and endotype 6-aminohexanoate-oligomer hydrolase (EIII) [11-14]. In Arthrobacter sp. KI72, enzymes EI, EII, and EIII are encoded from genes, nylA, nylB and nylC, respectively [11].
In the plasmid of Arthrobacter sp. KI723T1, all three genes (nylA, nylB and nylC) encoding enzymes with nylon 6 hydrolase activity were discovered [15]. These genes are also present in other nylon 6 degrading bacterial isolates belonging to a variety of taxa, including Pseudomonas, Agromyces, and Kocuria [16-18]. In the genome of Agromyces sp. KY5R, nylB and nylC are present, but it lacks nylA [17]. In Pseudomonas sp. NK87, in contrast, nylA and nylB are identified on the plasmids, but missing nylC [18].
The study of nylon 11 biodegradation is scarce. In a previous study by Kuo et al., nylon 11 was first treated with trifluoroacetic and blended with chitosan [19]. The authors demonstrated that the mass of chitosan-nylon 11 mixture decreased after being buried in soil for 10 to 40 days, indicating the possibility that nylon 11 is also biodegradable. However, to the best of our knowledge, the isolation of organisms that degrade nylon 11 or direct evidence of biodegradation by microorganisms has not been reported.
In our current study, to gain insights into a nylon 11 biodegradation process, we isolated a bacterial strain that grows on 11-aminoundecanoic acid, a precursor of nylon 11 synthesis and putative depolymerization product of nylon 11, from nylon 11 enrichment culture. There has been no previous report of the isolation of 11-aminoundecanoic acid metabolizing bacterial strains. The draft genome and phylogenetic analysis of the strain revealed that it belongs to genus Pseudomonas. The content of its genome included genes encoding extracellular hydrolases and homologues of nylon 6 hydrolases. Based on these findings, we postulated that a nylon 11 in enrichment culture was first cleaved by extra cellular enzyme(s) to form 11-aminoundecanoic acid, and then organisms such as JB-G utilized the 11-aminoundecanoic acid as carbon and nitrogen sources for growth.
Materials and Methods
Media and growth conditions
Cells were grown on defined media devoid of carbon and nitrogen by modifying basal salts medium (BSM) [20]. Specifically, ammonium chloride and nitrilotriacetic acid were omitted from the media (BSM-CN). Soil was collected from the Indiana University Kokomo campus. To construct nylon 11 enrichment culture, 1 g of soil and 1 g of nylon 11 pellets were added to 50 ml BSM-CN media. As a control, BSM-CN media containing succinate and nitrate as a carbon and nitrogen source was also created. After incubation at 30 °C with constant agitation at 150 rpm, cells were plated on BSM-CN agar medium supplemented with 11-aminoundecanoic acid as sole source of carbon and nitrogen.
For growth curve analysis, cells were transferred from overnight culture grown on BSM-CN media containing 11-aminoundecanoic acid (1%) to a fresh media, and grown at constant agitation at 150 rpm at 30 °C. The change in absorbance at 600 nm was monitored using the spectrophotometer. The growth was measured in triplicate.
Genome determination
The genomic DNA was purified using E.Z.N.A. Stool DNA kit (Omega Bio-tek, Inc., GA). Genome sequences were obtained by Illumina Hiseq 2500 sequencer at Macrogen Inc. (MD, USA). The de novo genome assembly was performed by Velvet 1.2.07 [21] and the graphical user interface Vague [22]. The K-mer size was chosen by software based on the genome size of 6 M bases. Genome annotation was conducted by RAST [23].
Phylogenetic analysis
Phylogenetic tree of 16S rRNA gene sequences was constructed via Maximum Likelihood method using MEGA6 with bootstrap values of 500 replications [24].
Results and Discussion
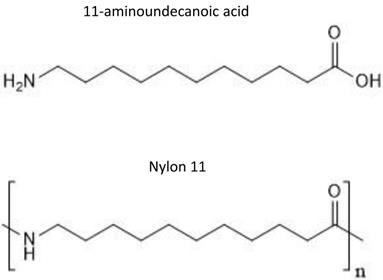
Strain JG-B was isolated from solid media containing 11-aminoundecanoic acid as a sole source of carbon and nitrogen. They were abundantly present on the solid media inoculated with nylon 11 enrichment culture but absent on the medium plated with cells from control enrichment culture. It also showed rapid growth on 11-aminoundecanoic acid in liquid culture (Figure 2). In order to rule out the possibility that the observed growth on 11-aminoundecanoic acid was due to the use of stored nitrogen in cells, growth in liquid culture was verified by repeated subculturing up to eight times. Atmospheric nitrogen was also a possible source of nitrogen for growth. Various growth tests were performed to eliminate this possibility. First, JG-B strain did not grow when carbon source alone (e.g. succinate) was added to the BSM-CN media (Table 1). Only when nitrogen was also provided as nitrate or as 11-aminoundecanoic acid, cells exhibited growth. Additionally, homologues of genes encoding known nitrogenase were not identified on the genome as described below.
Illumina sequencing of the genomic DNA produced a total 11.4 million reads which were de novo assembled into 90 contigs. The sequence has been deposited in NCBI GenBank under the accession number VOWX00000000. The total genome size is approximately 6 Mbp with 62.9 % GC content. It is composed of 5,367 coding sequences and 66 RNA genes (Table 2).
The phylogenetic analysis of the 16S rRNA gene sequences revealed that strain JG-B belongs to the genus Pseudomonas. Many Pseudomonas strains degrade a variety of aliphatic compounds [25, 26]. A phylogenetic tree was constructed with sequences of Pseudomonas strains with known biodegradation abilities (Figure 3). Notably, 16S rRNA gene sequence of JG-B strain showed a high sequence identity (97.5%) to that of P. aeruginosa PAO1. While wildtype strain PAO1 does not metabolize nylon 6, Prijambada et al. successfully created a mutant that degraded nylon 6 oligomers via experimental evolution [27].
Growth of Pseudomonas sp. strain JG-B with various carbon and nitrogen sources.
| Substrates | Carbon source | Nitrogen source | Growth |
|---|---|---|---|
| 11-aminoundecanoic acid | 11-aminoundecanoic acid | 11-aminoundecanoic acid | Growth |
| Succinate | Succinate | No | No growth |
| Succinate/nitrate | Succinate | Nitrate | Growth |
Genomic features of Pseudomonas sp. strain JG-B
| Features | |
|---|---|
| Total length (bp) | 5,991,756 |
| N50 | 279,576 |
| GC content (%) | 62.9 |
| Total number of genes | 5,588 |
| Protein coding gene (CDS) | 5,367 |
| rRNA genes | 3 |
| tRNA genes | 59 |
| ncRNA genes | 4 |
Growth curve of Pseudomonas sp. strain JG-B on 11-aminoundecanoic acid as sole source of carbon.
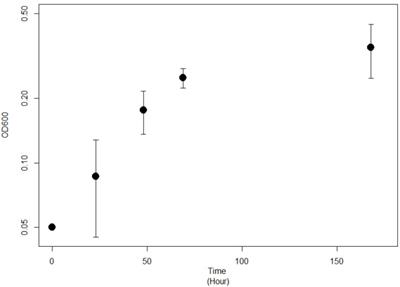
Phylogenetic tree of 16S rRNA gene sequences of Pseudomonas sp. strain JG-B and related strains. The tree was constructed using the maximum likelihood method. Numbers at nodes indicate bootstrap values. The scale bar represents nucleotide substitutions per site.
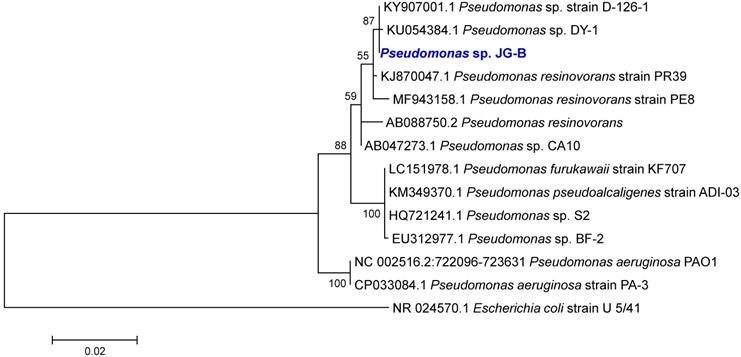
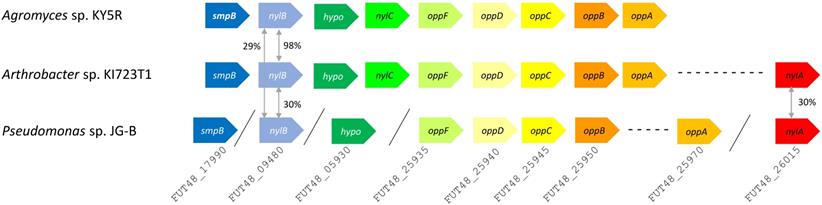
Putative nylon 11 degradation genes in Pseudomonas sp. strain JG-B and nylon 6 degrading bacteria. Abbreviations for genes and encoded proteins: smpB: tmRNA-binding protein; nylB: 6-aminohexanoate dimer hydrolase (EII); hypo: hypothetical protein; nylC: 6-aminohexanoate-oligomer hydrolase (EIII); oppF: oligopeptide transport ATP-binding protein; oppD: oligopeptide transport ATP-binding protein; oppC: oligopeptide transport system permease protein; oppB: oligopeptide transport system permease protein; oppA: periplasmic oligopeptide binding protein; and nylA: 6-aminohexanoate-cyclic-dimer hydrolase (EI). A symbol / indicates when genes are located on different contigs. The values represent protein sequence identity between the pair of orthologs.
Metabolism of 11-aminoundecanoic acid has never been reported. A recent study of the cellular metabolism of 6-aminohexanoate (the hydrolysis product of nylon 6) identified involvement of 5-aminovalerate aminotransferase/ Gamma-aminbutyrate:alpha-ketoglutarate aminotransferase (NylD) [28]. While 6-aminohexanoate and 11-aminoundecanoic acid differs significantly in the length of carbon chain (i.e. 6 vs 11), it is possible that it utilizes related enzymes and involves aminotransfer reaction. Blast analysis of NylD with JG-B showed that the JG-B genome contains a gene (FUT48_13600) which would encode a protein with 48 % protein sequence identity to NylD of Arthrobacter sp. KI72 (WP_079941468). Eleven additional genes encoding aminotransferase with a high similarity to Arthrobacter sp. KI72 NylD (i.e. >27% protein sequence identity) were also present on the strain JG-B's genome. Further research will be needed to decipher if any of these enzymes are involved in the 11-aminoundecanoic acid metabolism.
A variety of genes encoding hydrolases are also found on JG-B's genome, including a homologue of an extracellular lipase gene from Pseudomonas wisconsinensis (FUT48_16475), predicted extracellular lipase/esterase (FUT48_02000), and nylon 6 hydrolases (FUT48_26015 and FUT48_09480 encoding NylA and NylB, respectively) (Figure 4). NylA and NylB both shared 30 % protein sequence identity with corresponding proteins of Arthrobacter sp. KI723T1. A homologue of gene encoding NylC was absent on JG-B strain. In both Arthrobacter and Agromyces sequences, genes encoding subunits for oligopeptide permease (Opp) and tmRNA binding small protein B (SmpB) are present in the same region as genes encoding NylB and NylC. Both types of genes were also found on the genome of JG-B but they do not appear to be located in the neighboring region.
NylA is amidase, related to Asp-tRNAAsn/Glu-tRNAGln amidotransferase A subunit [29]. Through structural studies and mutagenesis of NylA in Arthrobacter, three residues at catalytic center (Ser174, Ser150 and Lys72) and a residue (Cys316), that are proposed to be involved in specific binding of NylA to its substrate 6-aminohexanoate cyclic dimer, were identified. Despite low sequence identity with Arthrobacter NylA (30%), JG-B's NylA contains identical catalytic residues, suggesting a conserved catalytic mechanism. In contrast, Cys316 is replaced by alanine in JG-B's NylA, suggesting different substrate specificity.
NylB is a penicillin-recognizing serine hydrolase sharing high structural similarity to DD-peptidase and carboxylesterase (EstB) [30]. In this penicillin recognizing family of serine hydroxylases, a serine residue of Ser-X-X-Lys motif functions as nucleophile in catalysis [30]. The motif was identified in the JG-B's NylB protein sequence. Like related NylB proteins, JG-B's NylB lacks other motifs present in EstB (e.g. Gly-X-Ser-X-Gly motif) and class A β-lactamases (e.g. KTG box). These observations suggest that catalytic residues appear to be located within the Ser-X-X-Lys motif.
Additionally, NylB from JG-B and Arthrobacter strain K172's EII enzyme share another active site residue (Y215 in 1WYB). However, residues that were shown to be important for 6-aminohexanoate linear dimer hydrolyzing activity were different or missing in JG-B's NylB sequence. Asp181 has no corresponding residue in JG-B's NylB, and Asn266 is replaced by Ser in JG-B [30]. These differences may reflect the divergences in their substrate(s).
In summary, we have isolated a Pseudomonas sp. strain JG-B that can grow on 11-aminoundecanoic acid as a sole source of carbon and nitrogen. Through genomic analysis, we identified genes that may be involved in the metabolism of 11-aminoundecanoic acid as well as its polymer nylon 11. We are currently investigating the nylon 11 degradation ability of this organism.
Acknowledgements
The study was funded by Grants-in Aid and Sustainability Grant from Indiana University Kokomo. J.G. was supported by undergraduate research scholarship. The authors also acknowledge the financial support from School of Sciences at Indiana University Kokomo. Authors thank Mr. Brandon Gatz-Schrupp for his critical reading of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mukhopadhyay SK. 8 - Manufacturing, properties and tensile failure of nylon fibres. Bunsell AR, editor. Handbook of Tensile Properties of Textile and Technical Fibres: Woodhead Publishing. 2009 p. 197-222
2. Lee B, Pometto AL, Fratzke A, Bailey TB. Biodegradation of Degradable Plastic Polyethylene by Phanerochaete and Streptomyces Species. Appl Environ Microbiol. 1991;57:678-85
3. Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW. et al. Biodegradation of polyester polyurethane by endophytic fungi. Appl Environ Microbiol. 2011;77:6076-84
4. Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y. et al. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science. 2016;351:1196-9
5. Deguchi T, Kakezawa M, Nishida T. Nylon biodegradation by lignin-degrading fungi. Appl Environ Microbiol. 1997;63:329-31
6. Kakudo S, Negoro S, Urabe I, Okada H. Nylon oligomer degradation gene, nylC, on plasmid pOAD2 from a Flavobacterium strain encodes endo-type 6-aminohexanoate oligomer hydrolase: purification and characterization of the nylC gene product. Appl Environ Microbiol. 1993;59:3978-80
7. Nakamura K, Tomita T, Abe N, Kamio Y. Purification and characterization of an extracellular poly(L-lactic acid) depolymerase from a soil isolate, Amycolatopsis sp. strain K104-1. Appl Environ Microbiol. 2001;67:345-53
8. Eberl A, Heumann S, Bruckner T, Araujo R, Cavaco-Paulo A, Kaufmann F. et al. Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules. J Biotechnol. 2009;143:207-12
9. Liebminger S, Eberl A, Sousa F, Heumann S, Fischer-Colbrie G, Cavaco-Paulo A. et al. Hydrolysis of PET and bis-(benzoyloxyethyl) terephthalate with a new polyesterase from Penicillium citrinum. Biocatal Biotransform. 2007;25:171-7
10. Tokiwa Y, Suzuki T. Hydrolysis of polyesters by lipases. Nature. 1977;270:76-8
11. Negoro S. Biodegradation of nylon oligomers. Appl Microbiol Biotechnol. 2000;54:461-6
12. Kinoshita S, Negoro S, Muramatsu M, Bisaria VS, Sawada S, Okada H. 6-Aminohexanoic Acid Cyclic Dimer Hydrolase. A New Cyclic Amide Hydrolase Produced by Acromobacter guttatus KI72. Eur J Biochem. 1977;80:489-95
13. Kinoshita S, Terada T, Taniguchi T, Takene Y, Masuda S, Matsunaga N. et al. Purification and characterization of 6-aminohexanoic-acid-oligomer hydrolase of Flavobacterium sp. Ki72. Eur J Biochem. 1981;116:547-51
14. Negoro S, Kakudo S, Urabe I, Okada H. A new nylon oligomer degradation gene (nylC) on plasmid pOAD2 from a Flavobacterium sp. J Bacteriol. 1992;174:7948-53
15. Kato K, Ohtsuki K, Koda Y, Maekawa T, Yomo T, Negoro S. et al. A plasmid encoding enzymes for nylon oligomer degradation: nucleotide sequence and analysis of pOAD2. Microbiology. 1995;141( Pt 10):2585-90
16. Yasuhira K, Tanaka Y, Shibata H, Kawashima Y, Ohara A, Kato D-i. et al. 6-Aminohexanoate Oligomer Hydrolases from the Alkalophilic Bacteria Agromyces sp. Strain KY5R and Kocuria sp. Strain KY2. Appl Environ Microbiol. 2007;73:7099-102
17. Yasuhira K, Uedo Y, Takeo M, Kato D, Negoro S. Genetic organization of nylon-oligomer-degrading enzymes from alkalophilic bacterium, Agromyces sp. KY5R. J Biosci Bioeng. 2007;104:521-4
18. Kanagawa K, Negoro S, Takada N, Okada H. Plasmid dependence of Pseudomonas sp. strain NK87 enzymes that degrade 6-aminohexanoate-cyclic dimer. J Bacteriol. 1989;171:3181-6
19. Kuo P-C, Sahu D, Yu HH. Properties and biodegradability of chitosan/nylon 11 blending films. Polym Degrad Stab. 2006;91:3097-102
20. Hareland WA, Crawford RL, Chapman PJ, Dagley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975;121:272-85
21. Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010 Chapter 11: Unit 11.5
22. Powell DR, Seemann T. VAGUE: a graphical user interface for the Velvet assembler. Bioinformatics. 2013;29:264-5
23. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA. et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC genomics. 2008;9:75
24. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725-9
25. Stanier RY, Palleroni NJ, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159-271
26. Cerniglia CE. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351-68
27. Prijambada ID, Negoro S, Yomo T, Urabe I. Emergence of nylon oligomer degradation enzymes in Pseudomonas aeruginosa PAO through experimental evolution. Appl Environ Microbiol. 1995;61:2020-2
28. Takehara I, Fujii T, Tanimoto Y, Kato DI, Takeo M, Negoro S. Metabolic pathway of 6-aminohexanoate in the nylon oligomer-degrading bacterium Arthrobacter sp. KI72: identification of the enzymes responsible for the conversion of 6-aminohexanoate to adipate. Appl Microbiol Biotechnol. 2018;102:801-14
29. Yasuhira K, Shibata N, Mongami G, Uedo Y, Atsumi Y, Kawashima Y. et al. X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation. J Biol Chem. 2010;285:1239-48
30. Negoro S, Ohki T, Shibata N, Mizuno N, Wakitani Y, Tsurukame J. et al. X-ray crystallographic analysis of 6-aminohexanoate-dimer hydrolase: molecular basis for the birth of a nylon oligomer-degrading enzyme. J Biol Chem. 2005;280:39644-52
Author contact
![]() Corresponding author: Hisako Masuda. School of Sciences, Indiana University Kokomo, Indiana 46902, United States of America.
Corresponding author: Hisako Masuda. School of Sciences, Indiana University Kokomo, Indiana 46902, United States of America.
Received 2019-11-14
Accepted 2020-1-2
Published 2020-1-25
